


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 31-12-2021
Date: 18-10-2021
Date: 13-12-2021
|
Polymerase Chain Reaction : Procedure
PCR uses DNA pol to repetitively amplify targeted portions of genomic or cDNA. Each cycle of amplification doubles the amount of DNA in the sample, leading to an exponential increase (2n, where n = cycle number) in DNA with repeated cycles of amplification. The amplified DNA products can then be separated by gel electrophoresis, detected by Southern blotting and hybridization, and sequenced.
1. Constructing primer:
It is not necessary to know the nucleotide sequence of the target DNA in the PCR method. However, it is necessary to know the nucleotide sequence of short segments on each side of the targe DNA. These stretches, called flanking sequences, bracket the DNA sequence of interest. The nucleotide sequences of the flanking regions are used to construct two, single-stranded oligonucleotides, usually 20–35 nucleotides long, which are complementary to the respective flanking sequences. The 3′-hydroxyl end of each oligonucleotide points toward the target sequence (see Fig. 1). These synthetic oligonucleotides function as primers in PCR.
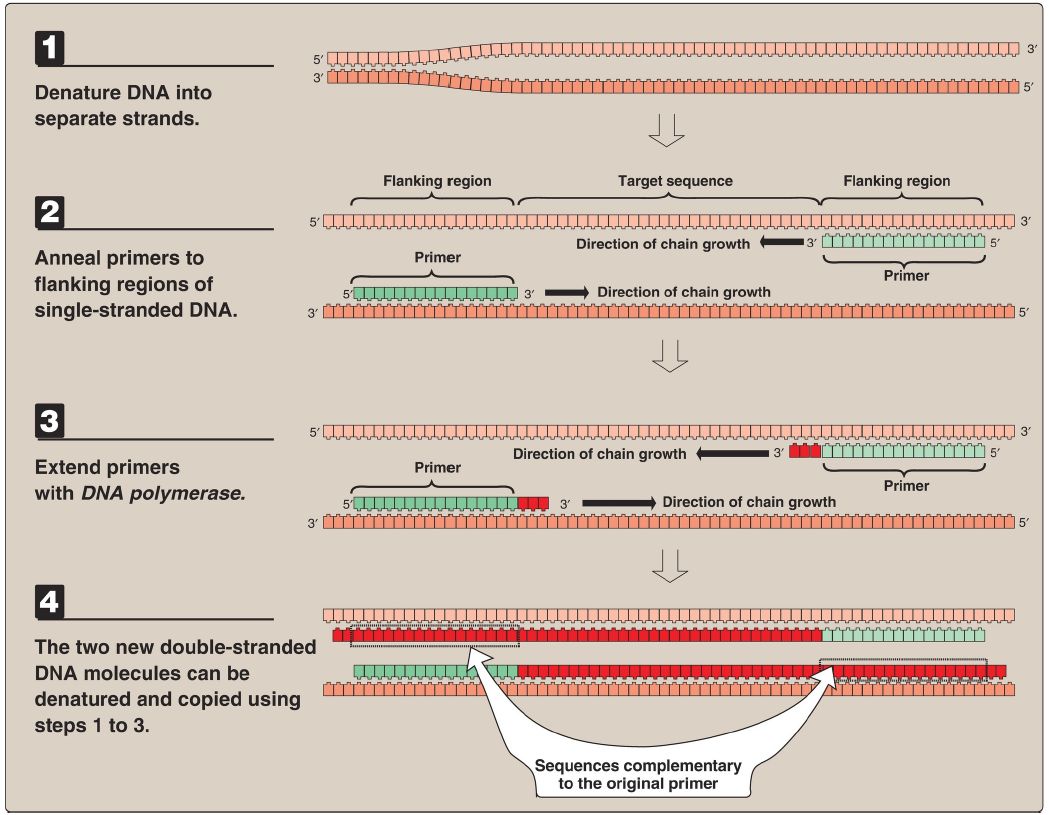
Figure 1: Steps (denature, anneal, extend) in one cycle of the polymerase chain reaction.
2. Denaturing DNA:
The target DNA to be amplified is heated to ~95°C to separate the dsDNA into single strands.
3. Annealing primers:
The separated strands are cooled to ~50°C and the two primers (one for each strand) anneal to a complementary sequence on the ssDNA.
4. Extending primers:
DNA pol and dNTP (in excess) are added to the mixture (~72°C) to initiate the synthesis of two new strands complementary to the original DNA strands. DNA pol adds nucleotides to the 3′-hydroxyl end of the primer, and strand growth extends in the 5′→3′ direction across the target DNA, making complementary copies of the target. [Note: PCR products can be several thousand base pairs long.] At the completion of one cycle of replication, the reaction mixture is heated again to separate the strands (of which there are now four). Each strand binds a complementary primer, and the step of primer extension is repeated. By using a heat-stable DNA pol (for example, Taq from the bacterium Thermus aquaticus that normally lives at high temperatures), the polymerase is not denatured and, therefore, does not have to be added at each successive cycle. However, Taq lacks proofreading activity.
Typically, 20–30 cycles are run during this process, amplifying the DNA by a million-fold (220) to a billion-fold (230). [Note: Each extension product includes a sequence at its 5′-end that is complementary to the primer (see Fig. 1). Thus, each newly synthesized strand can act as a template for the successive cycles (see Fig. 2). This leads to an exponential increase in the amount of target DNA with each cycle, hence, the name “polymerase chain reaction.”] Probes can be made during PCR by adding labeled nucleotides to the last few cycles.
Figure 1: Multiple cycles of the polymerase chain reaction.



|
|
|
|
فواكه قد تغني عن مضادات الحيوية وتظهر إمكانات غير متوقعة في مكافحة السرطان
|
|
|
|
|
|
|
الأكثر تطورا حتى الآن.. ميتا تكشف عن "إل لاما 4"
|
|
|
|
|
|
|
أصواتٌ قرآنية واعدة .. أكثر من 80 برعماً يشارك في المحفل القرآني الرمضاني بالصحن الحيدري الشريف
|
|
|