


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 27-10-2015
Date: 12-11-2015
Date: 11-11-2015
|
Molecular Basis of Inheritance
INTRODUCTION
The information regarding the expression of the genome is revealed by the analysis of chromosomes, and the DNA contained within them, using techniques at the molecular level. Chromosomes are the inherited elements through which the genetic material is transmitted. Within the chromosomes, the information-carrying component is DNA. Therefore the study of inheritance involves the study of the DNA sequence in genes. In order to understand genetic basis of a disease, and to use this information for diagnosis, possible treatment and the prevention, it is essential to know the structure and function of genes.
STRUCTURE OF A CHROMOSOME
The compact DNA forming a chromosome is composed of acidic chromosomal proteins called histones, and other heterogeneous proteins, non-histones. This DNA and protein complex is called chromatin. Histones are of five major types, and are termed as H1, H2A, H2B, H3 and H4, and they help in proper packaging of the chromatin. Two copies of these four histones form an octamer around which DNA winds. Each histone is associated with 140 base pairs, making two turns. Each DNA core complex is spaced by 20-60 base pairs. Thus the appearance of chromatin is like a beaded string. The complex of DNA and histones is called a nucleosome. Out of the five histones mentioned above, amino acid sequence of H1 varies more between species, while the other four show a conservation of amino acid sequences. The helical structure of the nucleosome is compacted into secondary chromatin structure, called solenoid. Under the electron microscope the chromatin structure appears three times thicker than the nucleosome fibre. Each turn of solenoid contains six nucleosomes. The solenoids are packed into loops, which are attached to non-histone proteins. The light and dark bands seen in prophase and metaphase chromosomes reflect the folding of clusters of loops, and also define functioning regions of the genome.
Structure of DNA
Deoxyribonucleic acid (DNA) or nucleic acid is the hereditary material, which is transmitted faithfully from parents to offspring during reproduction. First identified by F. Miescher in 1869, nucleic acids were first called “nuclein” because they were isolated from cell nuclei. In 1953, James Watson and Francis Crick on their X-ray diffraction studies, proposed a double helix model of DNA described below. For their work Watson and Crick, received a Noble Prize for Medicine and Physiology in 1962.
DNA is tightly bound coil, and lies in the condensed form within the nucleus of a cell in the form of rod shaped bodies called chromosomes (Fig.1). DNA is composed of repeating subunits called nucleotides. Each nucleotide is composed of a phosphate group, a five-carbon sugar (pentose), and a cyclic nitrogen-containing compound called a base. In DNA, the sugar is 2-deoxyribose. The DNA molecule consists of two complementary strands twisted in the form of a double helix. These complementary strands are chains composed of two types of nucleotide bases, pyrimidines and purines. The pyrimidine bases are thymine (T) and cytosine (C), and the purine bases are adenine (A) and guanine (G). The DNA model resembles a twisted ladder. The sides of the ladder form a backbone, and the links of DNA consist of deoxyribose residues linked by phosphate. The rungs of the ladder are made up of the bases purines and pyrimidines. Two strands of DNA are joined with each other by hydrogen bonds, which are present between the purine and pyrimidine bases. In a DNA molecule, adenine pairs with thymine (AT) and guanine with cytosine (GC). The ends of the DNA strands are designated as 5’ and 3’. The 5’ end is written to the left and indicates the sequence near the beginning of the gene, and 3’ is written to the right indicating the sequence near the end of the gene (Fig. 2).
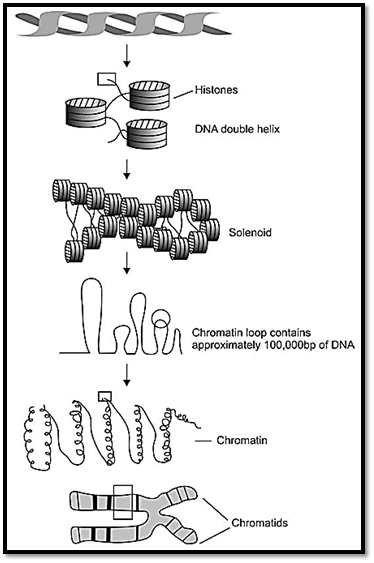
Fig. 1 : Structure of chromosome

Fig. 2: DNA molecule showing the sugar phosphate backbone and nucleotide pairing of A,G, C and T
THE GENETIC CODE
The genetic information is stored in the DNA molecule in a sequence of three bases, and this is referred to as the triplet code (Fig. 3). The coding unit, codon consists of three nucleotide bases each signifying an amino acid. For example, the codon UUC is the code for pheohis flexibility in third position in referred to as the wobble. The DNA code is said to be universal code and is same for all organisms, except is yeast mitochondria and mycoplasma. The complete ‘genetic code” was established by Nirenberg, Khorana and co-workers for which they were awarded Nobel prize in 1968 for physiology and medicine. The genetic message in the genetic code needs to be transmitted. The stages involved in this are replication, transcription and translation (Fig. 4).

Fig. 3: Genetic code

Fig. 4: Steps in the transfer of genetic information
Replication
In order to transmit genetic information from one generation to the next, the DNA molecule replicates. It is the process by which each strand of the parental DNA duplex is copied precisely by base pairing with complementary nucleotides. If the replication is errorless, the product is two duplexes identical in nucleotide sequence to the parental duplex. The genetic code is maintained during cell division. The new cell contains newly synthesized strands of DNA. This means, when a cell divides, genetic information encoded on the DNA is conserved and transmitted without change to daughter cells. The process is termed as semi-conservative replication. Two enzymes are required for this, DNA polymerase, and ligase. In a DNA molecule the replication starts at multiple points, which appears as a forked structure called the replication fork (Fig. 5). This occurs in both directions. Replication origins are usually 30300 kb apart, and each replication unit is 20-80 bp. Replication takes place during “S’ phase of the cycle until the total DNA is copied. Any damage that occurs in a DNA strand gets repaired and reconstituted the same way.

Fig. 5: Replication fork
Transcription
This is a process by which information contained in a DNA molecule is copied by base pairing, to form the complementary sequence of ribonucleotides. This is called messenger RNA (mRNA). The mRNA gets transported from nucleus to cytoplasm. Only 10% of DNA is transcribed into mRNA. The chemical difference between RNA and DNA is that sugar in RNA is ribose in place of deoxyribose in DNA, and the pyrimidine base is uracil (U) in RNA place of thymine (T). In retroviruses, the genetic material is in the form of RNA. This RNA is transcribed into DNA by the action of an enzyme called reverse transcriptase. This enzyme is of great practical value, as with the help of this DNA probes can be produced from RNA that corresponds to a coding sequence of a human gene. During transcription, the two strands of DNA separate in the area to be transcribed. One strand (the sense strand) functions as the template, and mRNA is formed under the influence of RNA polymerase. Transcription proceeds in a 5’ to 3’ direction. After some processing and modification, the mRNA molecule diffuses to the cytoplasm and the DNA strands reassociate.
Translation
Translation is a complex process that occurs on the ribosomes in the cytoplasm. In this step, information transcribed from DNA into messenger RNA (mRNA) directs the order of polymerisation of specific amino acids for the synthesis of proteins. Each mRNA molecule becomes attached to one or more ribosomes. As the ribosome moves along the RNA from the 5’ to the 3’ end, each codon is recognized by a matching transfer RNA (tRNA), which contributes its amino acid to the end of a new growing protein chain. Amino acids are successively added to the polypeptide chain till the stop codon is reached.
The structure of a gene
Introns and Exons
The sequences of most vertebrate genes are split into coding sequences called exons, which are separated by noncoding intervening sequences called introns. It was in 1977 that the intron-exon structure of genes was discovered. Introns are transcribed into the primary RNA transcript, but are spliced out of the mRNA before translation. A group of DNA sequences known as consensus sequences precisely control this process. Most eukaryotic genes are mainly composed of introns. The exact function of introns is not known but it is suggested that they increase the length of genes, so that reshuffling of genes between the homologous chromosomes during meiosis becomes easier. It is also suggested that introns may help to modify the time required for DNA replication. Exons are segments of the gene that remain after splicing of the primary RNA transcript (5’ untranslated sequences, coding sequences and 3’ untranslated sequences) (Fig. 6).
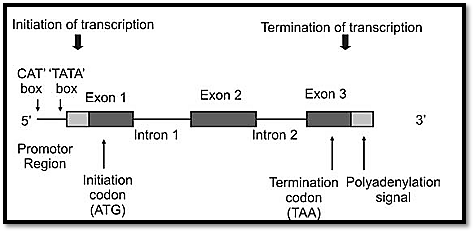
Fig. 6: Structure of a gene
Promoters and Enhancers
Promoters and enhancers are responsible for regulation of transcription. A promoter sequence is a combination of short sequence elements to which RNA polymerase binds in order to initiate transcription of a gene. Common promoter sequences are GC, TATA, CCAAT (these are called boxes). Transcription of genes starts in different places, thus producing different proteins. The same gene sequence is therefore known to cods for a variety of proteins in different tissues. Any mutation in the promoter sequence will therefore reduce the transcription level.
Another group of regulatory sequences are called enhancers. Enhancers are cis acting DNA sequences that can direct a significant increase in transcription, independent of their respective position and orientation within a given gene. Enhancers could be located thousands of base pairs away from promoters, but can interact with promoter sequences by a mechanism of DNA that allows multiple loop formation, which can permit interaction of many regulatory elements.
Extragenic repeated DNA sequences
The human nuclear genome contains a large amount of highly repeated DNA sequence families, which are largely transcriptionally inactive. A wide variety of different repeats are known. Noncoding repetitive DNA shows two major types of organization: tandemly repeated and interspersed.
Noncoding repetitive DNA
Tandemly Repeated Noncoding DNA
Such families are defined by blocks or arrays of tandemly repeated DNA sequences. Depending on the average size of the repeat units, highly repetitive noncoding DNA belonging to this class can be grouped into satellite (blocks from 100 kb to several Mb in length), minisatellite (blocks within the 0.1-20 kb) range and microsatellite DNA (blocks often less than 150 bp in range) (Fig. 7).
The major chromosomal location of satellite DNA is the centromeric chromatin of chromosomes and includes alphoid DNA and the Sau3A family. The major chromosomal location of the minisatellite DNA is at the telomeres and includes the hypervariable family and the telomeric family of minisatellite sequences.
Microsatellite sequences are dispersed throughout the human genome.
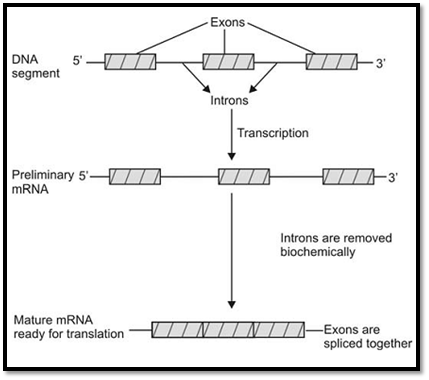
Fig. 7 : Removal of introns and splicing of exons during formation of a mature mRNA
Repetitive Interspersed DNA
Two major classes of interspersed repetitive DNA families have been discerned on the basis of repeat unit length, SINES (Short interspersed repeated sequences) and LINES (Long interspersed repeated sequences).
Short Interspersed Repeated Sequences (SINES)
The most conspicuous human SINE is the Alu repeat family (so called because of the early attempts at characterizing the sequence using the restriction endonuclease Alu I). The size of the repeat unit is ~0.3 kb, and there are about 1,000 000 copies present.
Long Interspersed Repeated Sequences (LINES)
Human LINES are exemplified by the LINE-1 or L-1 element. The size of the full length repeat unit is 6.1 kb, but the average size is ~0.8 kb. They are present at 200,000-500,000 copies per haploid genome.
Mutations
The term mutation refers to a sudden, heritable change in the genetic material of an organism or an individual. Such a newly formed phenotype is called a mutant. Mutants may result from faulty replication, movement, or repair of DNA and occur with a frequency of about one in every 106 cell divisions. Mutational changes in genetic material include changes in chromosomal number (aneuploidy, euploidy), structure, or in individual genes. Mutations can occur in the coding or non-coding regions of DNA molecule, and are of significance when they occur in the coding region. Mutations in the germ line may lead to inherited genetic diseases. If the mutation is dominant it will be passed on to half the germ cells, affecting half the progeny. If the mutation is recessive, it must be in the homozygous form to be expressed. When a child inherits identical mutations from both the parents he or she will become homozygous for that mutation. Mutations often affect somatic cells and so are passed on to successive generations cells within an organism. For example, most cancers are due to horizontal transmission of induced mutations, initially restricted to only that differentiated colony of cells. Mutations can occur spontaneously or may be induced by physical or chemical mutagens or by ionising radiations.
Importance of mutations
Mutation is the source of all genetic variation, and thus is the ultimate potential source for evolution. Alterations in alleles are mainly responsible for adaptation of individuals to the surrounding and for evolution in nature. On the other hand, mutations may be lethal to an individual, or may be responsible for disease.
Types of Mutations
Mutations can be divided into single base substitutions, which can be classified into silent, missense, and nonsense mutations. Other types of mutations include deletions, insertions, frameshift mutations, (which can be produced by deletions, insertions or splicing errors) and dynamic mutations, which include the triplet repeat mutations.
Single base Substitutions
Substitutions are the most common types of mutations, and as the name suggests it is the replacement of a single nucleotide by another. If a substituted nucleotide is replaced by the same type of nucleotide namely C for T or vice-versa, or A for G and vice-versa, it is called a transition. (a pyrimidine by pyrimidine or a purine by purine substitution). When a pyrimidine is substituted by a purine it is known as transversion. C to U transitions are more common and result in CpG dinucleotides. CpG dinucleotides get methylated in genomic DNA, with cytosine converting to thymine, and are called ‘hot spots’.
Silent Mutation
A mutation that does not alter the polypeptide product of the gene is termed as a synonymous or silent mutation. There is no alteration in the properties when a single base pair occurring in the third position of a codon results in another triplet coding for the same amino acid.
Missense Mutation
In a missense mutation, there is a single nucleotide substitution, which results in the coding for a different amino acid, resulting in the synthesis of an altered protein. The chemically dissimilar amino acid alters the protein structure. This is also known as non-conservative substitution, leading to a reduction or loss of biological function. Missense mutations lead to qualitative and not quantitative changes in protein function. So, even though the biological activity is maintained, there is a difference in the behaviour of the protein, an example of this being that of abnormal haemoglobins. The term conservative substitution is applied to a condition where chemically similar, but different amino acids are produced with no functional effect.
Nonsense Mutation
It occurs when a base pair substitution leads to the generation of a premature stop codon resulting in truncated proteins, and usually a dramatic reduction in gene function.
Deletions
A deletion involves the loss of one or more nucleotides. If the deletion occurs in a coding sequence and involves a single or multiple nucleotides, but not multiples of three, the reading frame is disrupted.
Insertions
An insertion can be an addition of one or more nucleotides in a particular gene. The situation is similar to a deletion whether it involves one or multiple nucleotides but not multiples of three, thus disrupting the reading frame.
Frameshift Mutation
Any mutation involving the insertion or deletion of one or a few nucleotides which are in the coding region, can change the triplet code. The reading frame being shifted, this constitutes a frameshift mutation.
Dynamic Mutations
Mutations can be passed unaltered to the next generation thus called stable mutations, or they may further alter during the process of transmission when they are called dynamic or unstable mutations. Triplet repeat expansions are included in the class of dynamic mutations.
In 1991, the genes for fragile X syndrome and spinobulbar muscular atrophy were found to contain unstable expanded trinucleotide repeats. This mechanism has now been implicated in several other diseases. The discovery of triplet repeat expansions finally allowed a molecular explanation for the inheritance pattern of anticipation. Anticipation is a phenomenon in which the age of onset of a disorder is reduced, and/or the severity of the phenotype is increased in successive generations. Anticipation has been observed in myotonic dystrophy, fragile X syndrome, Huntington disease, and autosomal dominant spinocerebellar ataxia, all of which are known to be caused by a similar mechanism.
Triplet repeats can be found in transcribed RNA destined to be untranslated (either 5’ or 3’ such as in fragile X syndrome or myotonic dystrophy respectively), spliced out intronic sequence (such as Freidreich ataxia) or coding exonic sequence (such as the dominant ataxias). In general non-coding repeats are able to undergo massive expansions from a normal number of 6-40 repeats to an abnormal range of many hundreds or thousands of repeats. This leads to either transcriptional suppression as in the case of fragile X syndrome or abnormal RNA processing limiting the amount of cytoplasmic message as in the case of myotonic dystrophy. In contrast, the coding expansions undergo much more modest expansions from a normal range of approximately 10-35 repeats to an abnormal range of approximately 40-90 repeats. Since these are CAG repeats coding for polyglutamine tracts, constraints of the individual protein structures significantly modify this range. CGG expansions occur in the 5’untranslated region in fragile X syndrome. Freidreich ataxia contains an intronic GAA repeat, and myotonic dystrophy contains a CTG expansion in the 3’ untranslated region. The expansion of CAG repeats within a coding segment of an exon is seen in Huntington’s disease, dentatorubral pallidoluysian atrophy (DRPLA), spinobulbar muscular atrophy (SBMA) and the spinocerebellar ataxias (SCAs). This results in proteins with elongated glutamine (Q) tracts.
Functional effects of mutations
Mutations of a gene might cause a phenotypic change in either of two ways:
1-The product may have reduced or no function, called a loss of function mutation. The alleles generated will be termed as null alleles (an allele that produces no effect) or hypomorphic alleles (an allele that produces a reduced amount or activity of a product)
2-The product may do something positively abnormal, called a gain of function mutation. The alleles generated will be termed as hypermorphic alleles (an allele that produces an increased amount or activity of the product) or neomorphic alleles (an allele with a novel activity or product).
Loss of Function Mutations
Loss of function mutations most often produces recessive phenotypes. For most gene products especially enzymes, the precise quantity is not crucial and we can get by on reduced levels of the product, up to half the normal amount. For some gene products however, 50% of the normal level is not sufficient for normal function. This is termed as haploinsufficiency and produces an abnormal phenotype, which is inherited in an autosomal dominant manner. Sometimes a non-functional mutant polypeptide interferes with the function of the normal allele in a heterozygous person. This is called the dominant negative effect.
Gain of Function Mutations
Gain of function mutations usually cause dominant phenotypes, because the presence of the normal allele does not prevent the mutant allele from behaving abnormally.
Mutagens
Mutations can arise spontaneously or due to environmental agents, which can alter the DNA or a chromosome. Such environmental agents are known as mutagens, and can be chemical, or ionising radiations (natural or artificial).
Chemical mutagens
There are 4 main groups of chemicals, which cause DNA mutations. Base analogues which mimic standard bases but pair improperly (5-bromouracil), alkylating agents which add alkyl groups to bases and so hamper correct pairing (nitrogen mustard), intercalating agents which intercalate with DNA and distort its structure (acroline dyes), and other agents which act directly on DNA (deamination by hydroxylamine).
Mustard gas, formaldehyde, and benzene are mutagenic in animals. Environmental chemicals that one can be exposed to, through use of pesticides in agriculture, industrial and pharmaceutical chemicals used commonly can be harmful. The Bhopal poison gas leakage in 1984 in India, is an example of a chemical mutagen, methyl isocynate which was responsible for chromosomal aberrations seen in affected survivors, and their offspring.
IONISING RADIATION
Ionizing radiation consists of short wave length X-rays, gamma rays and high-energy particles (alpha, beta and neutrons). X-rays, gamma rays and neutrons have high penetrating capacity but beta particles penetrate a few millimetres while the alpha particle penetrates only a fraction of millimetres, thus both penetrate only soft tissues. Heat, light and invisible radiation which man constantly receives from ground, air or from food and drink are also present. In the process of penetration, they produce ions by colliding with atoms of the material through which they penetrate and release electrons. The released electrons collide with other atoms releasing further electrons. The change in electron number, transforms a stable atom or molecule into a reactive ionic state. Thus along the tract of each high-energy ray, a train of reactive ions is formed, which can initiate a variety of chemical reactions affecting biological processes. Such an irradiation is called ionizing radiation.
The effect of radiation always depends on the dose of radiation and the measure used to calculate the radiation absorbed dose is called as rad. 1 rad measure denotes 100 ergs of energy actually absorbed by per gram of tissue exposed. Radiation for diagnostic purpose is a mixture of radiation measured as rem (roentgen equivalent for man). One rem is equivalent to 1 rad of plain X-ray. The amount of radiation received by an individual is measured in millisieverts (mSv), which is the commonly accepted unit. (100 Rem = 1Sv). Rem is the biological equivalent of Rad or Gray and is the accepted unit, and the term used in cases of medical exposure. [100 Rad=1Gray]. The critical dose in an adult is 500 Rads and more. However, for treatment of malignant illnesses, the dose far exceeds this, and may be as high as 5000-10000 Rads.
While studying mutations, exposure measurements in gonads are important as these effects are transmitted to progeny. These are of great significance in people who are exposed to X-rays as an occupation. Average human generation time is taken as 30 years approximately and gonadal dose of radiation is expressed as amount of rads in that period.
Natural sources of radiations
These include cosmic radiations from the earth's atmosphere, from the sun and the galaxy around it, the crust of the earth and rocks, the most important being Radon, which is emitted by rocks and can be trapped in buildings, and finally from our food and drink. The spontaneous rate of chromosomal breakage may be markedly increased due to exposure to ionising radiation or mutagenic chemicals. UV light causes formation of a pyrimidine dimmer in which pairs of adjacent pyrimidine bases become linked by carbon-carbon bonding. Dimerization produces a bulge in the affected DNA region, and cross links form interfering with DNA synthesis and RNA transcription. However there are naturally occurring DNA repair mechanisms and these mechanisms involve enzymes such as DNA glycosylases and nucleases.
Artificial sources of radiations
Doses from artificial sources of radiation are for most of the population much smaller than those from natural radiation. The artificial sources are the ones used for diagnostic and therapeutic purpose and those generated due to nuclear explosion.
Maximum Permissible Dose
The female oocyte is especially radiosensitive around the time of fertilization. An accidental diagnostic X-ray during the early stages of pregnancy results in the total added risk of 1 in 1000 to the fetes for congenital malformation, mental retardation or cancer. Neither termination of pregnancy or amniocentesis is indicated. Termination may be indicated if the fetes is less than 8 wks and the mother is exposed to 0.25 Gy or 25 rads. Exposure to 2-4 Gy results in female sterility. Exposure of either sex to therapeutic doses of radiation results initially in structural chromosomal abnormalities (rings, dicentrics, translocations) in 25-35% of lymphocytes. These tend to resolve by two years although translocations may persist.
The IAEA has recently updated its Basic Safety Standards together with the WHO, FAO and other such organisations. The occupational exposure permitted for a worker is 20 mSv per year. For the general public, the dose is 1 mSv per year, or in special circumstances up to 5 mSv in a single year provided the average over 5 years does not exceed 5mSv in 5 years. In the UK exposure limit is 15 mSv in a year. 1 mSv is approximately equal to 50 times the dose received in a single chest X-ray.
References
Purandarey , H. (2009) . Essentials of Human Genetics. Second Edition. Jaypee Brothers Medical Publishers (P) Ltd.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
المجمع العلمي ينظّم ندوة حوارية حول مفهوم العولمة الرقمية في بابل
|
|
|