


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 3-1-2016
Date: 16-3-2021
Date: 21-4-2016
|
The Basal Apparatus Assembles at the Promoter
KEY CONCEPTS
- The upstream elements and the factors that bind to them increase the frequency of initiation.
- Binding of TFII D to the TATA box or Inr is the first step in initiation.
- Other transcription factors bind to the complex in a defined order, extending the length of the protected region on DNA.
- When RNA polymerase II binds to the complex, it may initiate transcription.
In a cell, gene promoters can be found in three basic types of chromatin with respect to activity. The first is an inactive gene in closed chromatin. The second is a potentially active gene in open chromatin bound with RNA polymerase, called a poised gene.
Promoters in this class may assemble the basal apparatus, but they cannot proceed to transcribe the gene without a second signal to start transcription. Heat-shock genes are poised so that they
can be activated immediately upon a rise in temperature. The third class (which we will examine shortly) is a gene being turned on in open chromatin.
What has been largely unexplored until recently is the involvement of noncoding RNA (ncRNA) transcripts in gene activation. Numerous recent examples have been described in which transcription of ncRNAs regulates transcription of nearby or overlapping protein-coding genes. The production of these functional ncRNAs (also referred to as cryptic unstable transcripts, or CUTs) is much more common than originally believed. A significant number of active promoters have transcripts generated upstream of the promoters (known as promoter upstream transcripts, or PROMPTs). PROMPTs are transcribed in both sense and antisense orientations relative to the downstream promoter and may play a regulatory role in transcription. The many roles of ncRNAs in transcriptional regulation are discussed further in the chapter titled Regulatory RNA.
The initiation process requires the basal transcription factors to act in a defined order to build a complex that will be joined by RNA polymerase. The series of events summarized in FIGURE 1 is one model. It is important to remember that RNA polymerase II promoters are structurally very diverse. Once a polymerase is bound, its activity then is controlled by enhancer-binding transcription factors.
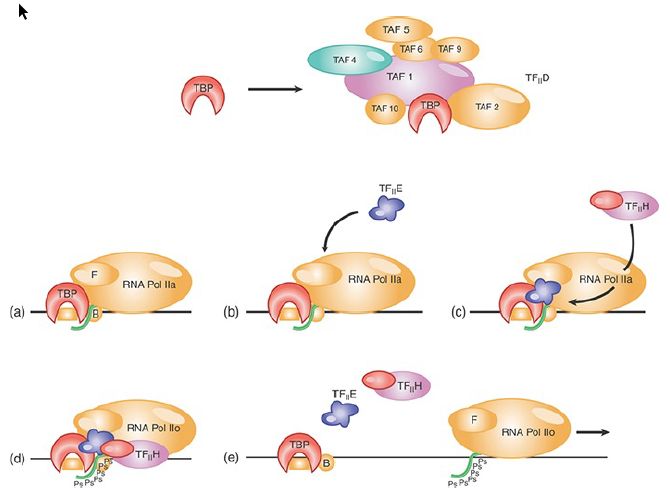
FIGURE 1. An initiation complex assembles at promoters for RNA polymerase II by an ordered sequence of association with transcription factors. TFII D consists of TBP plus its associated TAFs as shown in the top panel; TBP alone, rather than TFII D, is shown in the remaining panels for simplicity.
Data from M. E. Maxon, J. A. Goodrich, and R. Tijan, Genes Dev. 8 (1994): 515–524.
A promoter for RNA polymerase II often consists of two types of regions. The core promoter contains the start point itself, typically identified by the Inr, and often includes either a nearby TATA box or DPE; additional less common elements may be found as well. The efficiency and specificity with which a promoter is recognized, however, depend upon short sequences farther upstream, which are recognized by a different group of transcription factors, sometimes called activators. In general, the target sequences are about 100 bp upstream of the start point, but sometimes they are more distant. Binding of activators at these sites may influence the formation of the initiation complex at (probably) any one of several stages. Promoters are organized on a principle of “mix and match.” A variety of elements can contribute to promoter function, but none is essential for all promoters.
The first step in activating a TATA box–containing promoter in open chromatin is initiated when the TBP subunit of TFII D directs its binding to the TATA box. This may be enhanced by upstream elements acting through a coactivator. (TFII D also recognizes theInr sequence at the start point, the DPE, and other promoter elements.) TFII B binds downstream of the TATA box, adjacent to TBP in a region called the B recognition element (BRE), thus extending contacts along one face of the DNA from −10 to +10. The crystal structure of the ternary complex shown in FIGURE 2 extends this model. TFII B makes contacts in the minor groove downstream of the TATA box, and contacts the major groove upstream of the TATA box. In archaeans, the homolog of TFII B actually makes sequence-specific contacts with the promoter in the BRE region. This step is believed to be the major determinant in the establishment of promoter polarity, which way the RNA polymerase faces, and thus which strand is the template strand. TFII B may provide the surface that is, in turn, recognized by RNA polymerase, so that it is responsible for the directionality of the polymerase binding. TFII B also has a major role in recruiting RNA pol II to the TFII D/TF A/promoter DNA complex, assisting in the conversion from the closed to the open complex, and selecting the transcription start site (TSS).
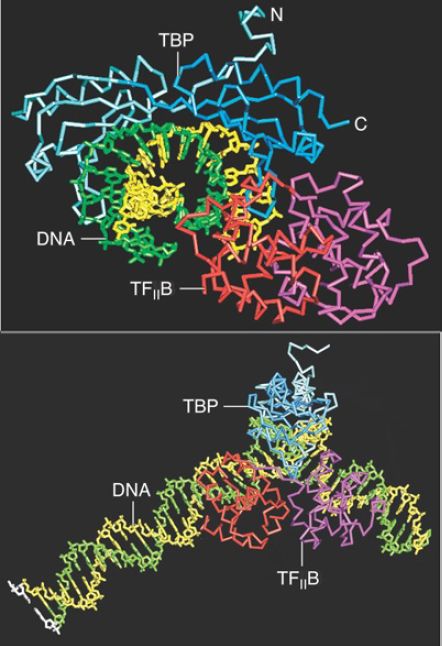
FIGURE 2. Two views of the ternary complex of TF B-TBPDNA show that TFII B binds along the bent face of DNA. The two strands of DNA are green and yellow, TBP is blue, and TF B is red and purple.
Photo courtesy of Stephen K. Burley.
The crystal structure of TFII B with RNA polymerase shows that three domains of the factor interact with the enzyme. As illustrated schematically in FIGURE 3, an N-terminal zinc ribbon from TFII B contacts the enzyme near the site where RNA exits; it is possible that this interferes with the exit of RNA and influences the switch from abortive initiation to promoter escape. An elongated “finger” of TFII B is inserted into the polymerase active center. The C-terminal domain interacts with the RNA polymerase and with TFIID to stabilize initial promoter melting. It also determines the path of the DNA where it contacts the factors TFII E, TFII F, and TFII H, which may align them in the basal factor complex.
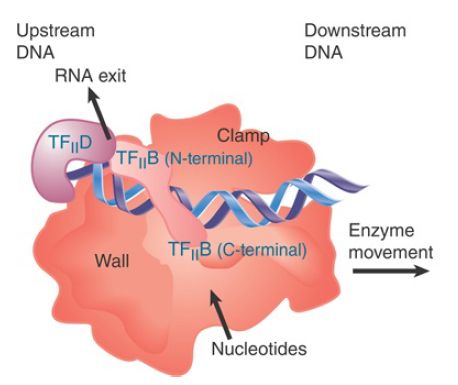
FIGURE 3. TFII B binds to DNA and contacts RNA polymerase near the RNA exit site and at the active center, and orients it on DNA. Compare with Figure 2, which shows the polymerase structure engaged in transcription.
The factor TFII F is a heterotetramer consisting of two types of subunits and is required for PIC (preinitiation complex) assembly. The larger subunit (RAP74) has an ATP-dependent DNA helicase activity that could be involved in melting the DNA at initiation. The smaller subunit (RAP38) has some homology to the regions of bacterial sigma factor that contact the core polymerase; it binds tightly to RNA polymerase II. TFII F may assist in bringing RNA polymerase II to the assembling transcription complex and is required, along with TF B, for transcription start-site selection. The complex of TBP and TAFs may interact with the CTD tail of RNA polymerase, and interaction with TF B may also be important when TFII F/polymerase joins the complex. Polymerase binding extends the sites that are protected downstream to +15 on the template strand and +20 on the nontemplate strand. The enzyme extends the full length of the complex because additional protection is seen at the upstream boundary.
What happens at TATA-less promoters? The same general transcription factors, including TF D, are needed. The Inr provides the positioning element; TFII D binds to it via an ability of one or more of the TAFs to recognize the Inr directly. Other TAFs in TFII D also recognize the DPE element downstream from the start point. The function of TBP at these promoters is more like that at promoters for RNA polymerase I and at internal promoters for RNA polymerase III.
When a TATA box is present, it determines the location of the start point. Its deletion causes the site of initiation to become erratic, although any overall reduction in transcription is relatively small.
Indeed, some TATA-less promoters lack unique start points, so initiation occurs within a cluster of start points. The TATA box aligns the RNA polymerase via the interaction with TFII D and other factors so that it initiates at the proper site. Binding of TBP to TATA is the predominant feature in recognition of the promoter, but two large TAFs (TAF1 and TAF2) also contact DNA in the vicinity of the start point and influence the efficiency of the reaction. Whereas most of the genes that RNA polymerase II transcribes are protein-coding mRNA genes, RNA pol II also transcribes some of the minor class snRNA genes. These have a similar, but not identical, promoter. Transcription of snRNA and the snoRNA (small nucleolar) genes in the nucleolus requires a specific modification of the CTD, a specific methylation of an Arg residue.
Assembly of the RNA polymerase II initiation complex provides an interesting contrast with prokaryotic transcription. Bacterial RNA polymerase is essentially a coherent aggregate with intrinsic ability to recognize and bind the promoter DNA; the sigma factor, needed
for initiation but not for elongation, becomes part of the enzyme before DNA is bound, although it may later be released. RNA polymerase II can bind to the promoter, but only after separate transcription factors have bound. The transcription factors play a role analogous to that of bacterial sigma factor—to allow the basic polymerase to recognize DNA specifically at promoter sequences— but have evolved more independence. Indeed, the factors are primarily responsible for the specificity of promoter recognition.
Only some of the factors participate in protein–DNA contacts (and only TBP and certain TAFs make sequence-specific contacts); thus protein–protein interactions are important in the assembly of the complex.
Although assembly can take place just at the core promoter in vitro, this reaction is not sufficient for transcription in vivo, where interactions with activators that recognize the more upstream elements are required. The activators interact with the basal apparatus at various stages during its assembly .



|
|
|
|
دخلت غرفة فنسيت ماذا تريد من داخلها.. خبير يفسر الحالة
|
|
|
|
|
|
|
ثورة طبية.. ابتكار أصغر جهاز لتنظيم ضربات القلب في العالم
|
|
|
|
|
|
|
سماحة السيد الصافي يؤكد ضرورة تعريف المجتمعات بأهمية مبادئ أهل البيت (عليهم السلام) في إيجاد حلول للمشاكل الاجتماعية
|
|
|