


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 30-12-2015
Date: 7-6-2021
Date: 29-11-2015
|
Translation Can Be Regulated
KEY CONCEPTS
-Translation can be regulated by the 5′ untranslated region (UTR) of the mRNA.
-Translation may be regulated by the abundance of various tRNAs.
- A repressor protein can regulate translation by preventing a ribosome from binding to an initiation codon.
- Accessibility of initiation codons in a polycistronic mRNA can be controlled by changes in the structure of the mRNA that occur as the result of translation.
Control over which and how much protein is made occurs first at the level of transcription control ; then through RNA-processing control (rare in bacteria, but common in eukaryotes); and, finally, translation-level control, which is examined here. (Refer to The Operon chapter for detail on the lac operon and its regulation.)
The lac repressor is encoded by the lacI gene; this is an unregulated gene that is continuously transcribed, but from a poor promoter. Also, the coding region of the lac repressor is in a very “poor” mRNA, meaning that the 5′ UTR of the mRNA has a poor sequence context that does not allow rapid ribosome binding or movement onto the ORF. Just as promoters can be “good” or “poor,” so can mRNAs. Together, this means that ribosomes do not translate the small amount of mRNA at the same level as the lacZYA polycistronic mRNA. Thus, very little lac repressor is found in a cell—only about 10 tetramers.
A second way that translation can be modulated is by codon usage. Multiple codons exist for most of the amino acids. These codons are not utilized equally by tRNAs; some have abundant
tRNAs, others do not. An ORF consisting of codons with abundant tRNAs can be rapidly translated, whereas another ORF that contains codons with less-abundant tRNAs will be translated more slowly.
Additionally, more active mechanisms exist for translation-level control. One mechanism for controlling gene expression at the level of translation parallels the use of a repressor to prevent
transcription. Translational repression occurs when a protein binds to a target region on mRNA to prevent ribosomes from recognizing the initiation region. Formally, protein–mRNA binding is equivalent to a repressor protein binding to DNA to prevent polymerase from utilizing a promoter. Polycistronic RNA allows coordinate regulation of translation, analogous to transcription repression of an operon. Figure 1 illustrates the most common form of this interaction, in which the regulator protein binds directly to a sequence that includes the AUG initiation codon, thereby preventing the ribosome from binding.

FIGURE 1. A regulator protein may block translation by binding to a site on mRNA that overlaps the ribosome-binding site at the initiation codon.
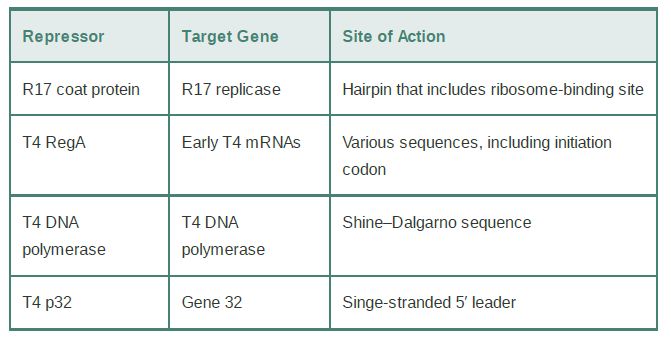
Some examples of translational repressors and their targets are summarized in Table 1. A classic example of how the product of translation can directly control the translation of its mRNA is the coat protein of the RNA phage R17; it binds to a hairpin that encompasses the ribosome-binding site in the phage mRNA. Similarly, the phage T4 RegA protein binds to a consensus sequence that includes the AUG initiation codon in several T4 early mRNAs, and T4 DNA polymerase binds to a sequence in its own mRNA that includes the Shine–Dalgarno element needed for ribosome binding.
TABLE 1.Proteins that bind to sequences within the initiation regions of mRNAs may function as translational repressors.
Another form of translational control occurs when translation of one gene requires changes in secondary structure that depend on translation of an immediately preceding gene. This happens during translation of the RNA phages, whose genes always are expressed in a set order. Figure 2 shows that the phage RNA takes up a secondary structure in which only one initiation sequence is accessible; the second cannot be recognized by ribosomes because it is base paired with other regions of the RNA. However, translation of the first gene disrupts the secondary structure, allowing ribosomes to bind to the initiation site of the next gene. In this mRNA, secondary structure controls translatability.
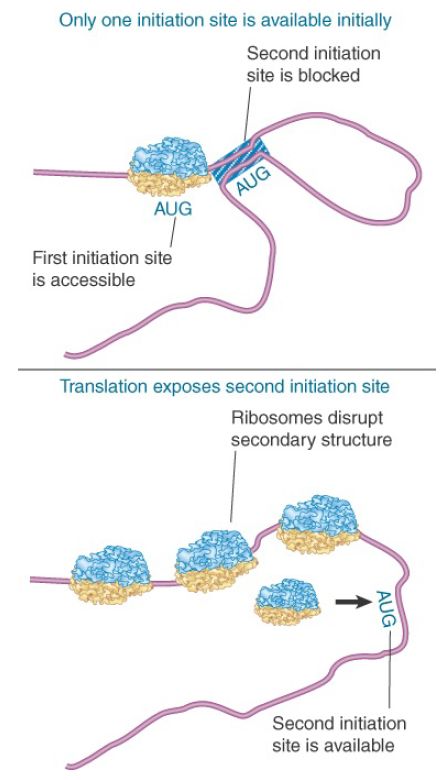
FIGURE 2. Secondary structure can control initiation. Only one initiation site is available in the RNA phage, but translation of the first gene changes the conformation of the RNA so that other initiation site(s) become available.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
اتحاد كليات الطب الملكية البريطانية يشيد بالمستوى العلمي لطلبة جامعة العميد وبيئتها التعليمية
|
|
|