


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 13-12-2015
Date: 28-4-2016
Date: 5-5-2021
|
Apoptosis
Wyllie et al. initially used the term “apoptosis” in a paper in 1972 to describe a newly observed form of cell death (1). The discovery of apoptosis has revolutionized many areas of modern biology, especially those areas concerned with disease development (2). This is primarily because apoptosis is an active form of cell death that, unlike necrosis, is critical for the maintenance of tissue homeostasis (3, 4). Thus, cell populations are numerically controlled through not only differentiation and proliferation, but also the physiological loss of cells through apoptosis. Critically, this implies that a cell which loses the capacity to apoptose, or a cell which becomes inappropriately sensitive to death stimuli, can lead to perturbations in cell number regulation and, ultimately, disease development.
Cells undergoing apoptosis are evident in all tissues in response to diverse stimuli. In healthy tissues, apoptosis accounts for all the cell deaths occurring in response to normal physiological signals. Apoptosis (often referred to as programmed cell death in a developmental context) occurs at many stages of embryonic development, eg, during the formation of digits from a solid limb paddle, where interdigital cells between the digits die (5). Single apoptotic cells are evident in healthy proliferating tissues such as small gut crypts and the dermis of the skin (4). Moreover, apoptosis is also seen during endocrine-induced atrophy of tissues, in cell-mediated immunity, in development of the nervous system, and in the “necrotic” oxygen-starved cores of solid tumors (6).
Morphologically, an apoptotic cell is very different from a necrotic cell (1, 6, 7); see Figure 1. One of the most prominent and easily identifiable stages of apoptosis involves the nucleus. Chromatin condenses and forms aggregates near the nuclear membrane, which becomes convoluted, whereas the nucleolus becomes enlarged and appears abnormally granular. The chromatin is also subject to the actions of an activated endonuclease that cleaves the DNA into 300- to 50-kbp fragments initially, and 180-bp fragments subsequently (8). Changes in the cytoplasm are also evident at this time; the cell shrinks visibly, adherent cells round up, and distinct protuberances or membrane blebs are discernible. Organelles within the shrunken cytoplasm still look normal, except for dilation of the endoplasmic reticulum. These “blebbing” cells exclude vital dyes, indicating no structural failure in the cell membrane. It is at this point in the apoptotic process, which may only take 10 to 30 minutes, that apoptotic cells in vivo are phagocytosed, by either their nearest neighbors or professional macrophages (9). Thus, apoptosis occurring in single cells in vivo is very easily overlooked due to its rapid progression. Cells in the later stages of apoptosis, especially those in vitro, form apoptotic bodies as a result of pinching off of the highly convoluted blebbing areas of the cell. These apoptotic bodies are phagocytosed or, in situations where there is much cell death, the unphagocytosed apoptotic cells undergo secondary necrosis, characteristically swelling and loosing membrane integrity.

Figure 1. A comparison of apoptosis and necrosis.
1. Removal of Apoptotic Cells
A critical part of the apoptotic pathway is the efficient recognition and phagocytosis of apoptotic cells. The rapid disposal of apoptotic cells does not elicit an immune response, consistent with apoptosis being the “no-nonsense” pathway for the disposal of unwanted cells. A breakdown in either the communication between macrophage and apoptotic cell or in the pathway of apoptosis itself may be two of the mechanisms through which chronic inflammatory disorders occur. Recognition of apoptotic cells by professional macrophages is mediated partly by avb3 integrin (the vitronectin receptor) and the CD36 ligand. Between these two molecules on the external surface of the cell sits a glycoprotein containing the sequence -R-G-D- (-Arg-Gly-Asp-), thrombospondin, which acts as a molecular bridge to bind to an unknown anionic site on the apoptotic cell (10-12.( Apoptotic cells also express surface phosphatidylserine, which binds to an as yet uncharacterized macrophage receptor (13, 14). The recognition of apoptotic cells mediated by the vitronectin receptor, CD36 ligand, and thrombospondin is confined to monocyte-derived macrophages. Recognition of apoptotic cells through the expression of phosphatidylserine, which is flipped onto the cellular surface due to the inactivation of an ATP-dependent flipase, is utilized primarily by inflammatory macrophages (9, 15). Four other molecules have been implicated in apoptotic cell recognition systems: (1) the 61D3-antigen found on human macrophages, (2) ICAM-3, found on the surface of human B cells (16), (3) the ABC1 transporter (17), and (4) the macrophage scavenger receptor (18). As mentioned previously, the interaction between apoptotic cells and macrophages does not elicit an inflammatory response, unless the apoptotic cells start to undergo secondary necrosis. Under these circumstances, other macrophage receptors are employed, and inflammatory cytokines are released (19). This change from “silent” death to one that activates the immune system may be a method of recruiting more phagocytes to the site of cell death to cope with the increasing number of corpses.
2. Regulation of Apoptosis and Cell Viability
The discovery that apoptosis is an active form of cell death which is transiently suppressable by both inhibitors of protein and RNA synthesis suggested that the cell needed to synthesize new proteins prior to its death [reviewed in (20)]. This sparked an enthusiastic hunt for a critical cell death gene that had to be translated prior to death. No such gene has been found, but many genes that regulate apoptosis have been identified as a result.
A cell can be triggered to undergo apoptosis in response to diverse stimuli, with each stimulus activating different pathways involving gene expression, as well as post-translational modification. The discovery that apoptosis can be suppressed by the presence of specific survival factors, either cytokines or proteins expressed within the cell, suggested that regulation of cell viability was paramount to the function of the cell (21). Indeed, it appears that apoptosis represents a default pathway in all cells (22). If a cell does not receive the correct survival stimulus, it will die; consequently, cells within multicellular organisms are maintained in a viable state by a constant supply of survival factors.
Once a cell is triggered to apoptose, activation of a common set of destruction proteins, irrespective of the stimulus involved, precipitates the morphological changes that we call apoptosis. Overall, the apoptotic pathway can be divided into three phases:
1. The decision phase, in which the cell receives a stimulus and, depending on both its internal and external environments, may or may not be triggered to die.
2. The commitment phase, in which the cell is committed to death and cannot recover.
3. The execution phase, in which the decision to die has been made and the activated apoptotic machinery leads to the morphological changes that define an apoptotic cell.
3. The Decision Phase of Apoptosis
The decision phase can be compared to that of a judge hearing the evidence during a trial. The evidence presented comes from many different sources: genes, cytokines, toxic chemicals, DNA damage, and the presence of viral and bacterial infection. Only once all the evidence is heard will the sentence be passed. The genes that modulate the decision phase are many and varied. Some are more commonly associated with the regulation of the cell cycle, such as c-myc, cdc25, c-fos, p53 and rb )retinoblastoma), whereas others are members of recently identified gene families, such as the bcl-2 family. Cytokines can either trigger or delay apoptosis. Tumor necrosis factor a (TNFa) and CD95 ligand (CD95L, FasL, Apo-1L) are two examples of cytokines that trigger apoptosis upon binding to their receptors (23), whereas the addition of factors such as interleukin-3 (IL-3) (24) or insulin-like growth factor 1 (IGF-1) suppresses death (25). Conversely, removal of anti-apoptotic cytokines also induces cell death (26, 27). Both viruses and bacteria trigger apoptosis upon infection of a cell, suggesting that the cell commits suicide to prevent the invading pathogen from spreading. However, both viruses and bacteria have evolved mechanisms that manipulate this response. Adenovirus, eg, has three genes required for productive infection: E1A, E1Bp19, and p55. E1A stimulates the infected cell to proliferate; however, this stimulus concomitantly triggers apoptosis. E1Bp19 and p55 delay the host cell from triggering cell death, thereby facilitating virus production (28, 29).
Alternatively, some bacteria such as Shigella flexneri actively promote the death of the cell by triggering the execution machinery (30-32). This allows the early release of the bacteria and triggers an immune response, which damages the surrounding tissue and aids the passage of the bacteria into its target cells.
3.1. Oncogenes, Tumor Suppressor Genes, and Apoptosis
In terms of suppressing tumorigenesis, long-lived organisms like humans need to limit the chances of their component cells acquiring mutations that lead to increased proliferative capacity and eventual clonogenic outgrowth. This is, in part, limited by restricting the types of cells with proliferative capacity. Many adult cell lineages exist in a senescent or postreplicative state. Moreover, for those cells that must retain the capacity to divide, apoptosis provides a “safety net.” Genes that are involved in proliferation are also involved in cell death; in fact, the two processes appear inexorably linked. This paradoxical coupling of two opposing biological processes was first realized through work on the pervasive oncogene c-myc (33) but has since been confirmed for several mitotic genes (34).
3.2. c-myc and Apoptosis
The protein product of the c-myc gene, c-Myc, is both necessary and sufficient to ensure fibroblast proliferation; in the presence of mitogen, all cells in cycle express c-Myc. Fibroblasts remain in cycle as long as mitogens and c-Myc are present, but only remain viable in the presence of additional survival factors. In the absence of such survival factors, cycling cells expressing deregulated Myc undergo apoptosis. This response to oncogene deregulation is thought to protect the organism against cells that acquire mutations in c-Myc. Mutant cells that continuously express c-Myc will proliferate, but since the survival factor in vivo is thought to be limiting, the mutant cells will die once the supply of factor is exhausted (25, 35). Thus, small proto-tumors may well arise in vivo, but they should regress due to lack of survival signals. If, however, another mutation has occurred producing a cooperating anti-apoptotic lesion, the mutated cells will survive. Several genes that cooperate with myc during tumorigenesis are anti-apoptotic. For example, v-Abl, which is a very good suppressor of apoptosis in response to many stimuli, will render haemopoietic cells cytokine-independent for growth and survival (36, 37). bcl-2, a gene initially identified through its translocation in the t(14:18( mutation found in follicular lymphoma (38), will cooperate with myc, suppressing the apoptotic signal induced through Myc expression (39, 40). The outcome of this cooperation in vivo is lymphoma, as seen in both double bcl-2/Eµ-myc transgenic mice (41, 42) and in human patients with the t(14:18) translocation (43, 44).
3.3. Bcl-2, an Anti-apoptotic Oncogene
The potent ability of Bcl-2 to suppress apoptosis triggered in response to a number of diverse stimuli prompted much research into its biochemical function. This function is still relatively obscure, but several members of the Bcl-2 family of proteins have been identified as a result (45-47). Bcl-2's form and functionality have been conserved throughout multicellular evolution, as exemplified by Ced-9, its counterpart in the nematode C. elegans, and the Adenovirus E1Bp19 protein (48). Ten or so members of this family now exist, and all share defined regions of homology, although not all act to suppress apoptosis (Table 1). The pro-apoptotic members of the family (Bax, Bak, and Bcl-xS (49-53)) share three regions of homology, BH-1, -2, -3, whereas the anti-apoptotic members (Bcl-2 and Bcl-xL) have a fourth region of homology, known as BH-4 (54, 55). Removal of the BH-4 domain from either Bcl-2 or Bcl-xL results in a loss of protective function. Family members can dimerize with themselves and with one another, and these interactions are important for their function (56( 57, . For example, Bcl-2 preferentially binds to the pro-apoptotic family member Bax, whereas Bcl-xL binds to Bak. If the number of Bax homodimers in the cell exceeds the number of Bcl-2 homodimers or Bax/Bcl-2 heterodimers, then the cell is more likely to undergo apoptosis (47). However, it is not clear which forms of these proteins are dominant for determining life or death. A second set of Bcl-2-associated proteins has been identified that influences cell survival through interaction with Bcl-2 family members, but whose members do not have all, if any, of the BH regions (Table 2) (45, 46). Bag-1, eg, has no homology to Bcl-2, but binds to it and augments the protective function of Bcl-2 (58). Bad, on the other hand, has homology to the family and binds Bcl-2, an interaction that disrupts Bcl-2's binding with Bax, leading to cell death (59, 60).
Table 1. Pro-apoptotic and Anti-apoptotic Members of the Bcl-2 Familya
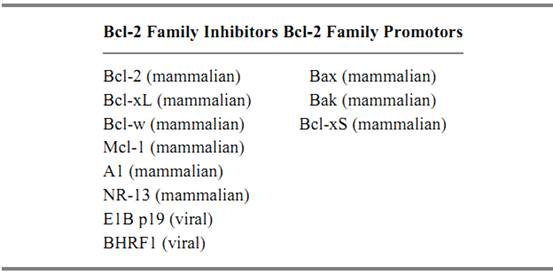
a Anti-apoptotic members contain up to 4 Bcl-2 homology (BH) domains and the transmembrane (TM) domain, whereas pro-apoptotic members contain only BH 1-3 and the TM domain, except for Bcl-xs that contains only BH4, BH3, and the TM domain. Members of this gene family have been conserved throughout evolution with homologues existing in nematodes, mammals, and viruses (46, 47, and references therein].
Table 2. Proteins That Bind to Bcl-2 Family Members and Regulate Their Function a
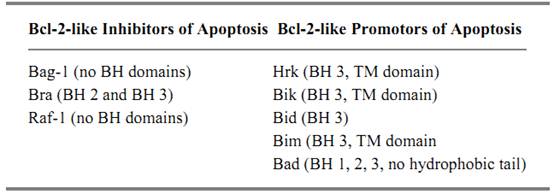
a A variety of proteins interact with Bcl-2 proteins. Some contain the BH domains that influence their function, such as Bid, Bim, Bik, whereas others have no homology with Bcl-2, but still bind and influence its function (46, 47, and references therein].
Both Bcl-xL and Bcl-2 have effects on cell-cycle transition (61). Cells expressing either gene exhibit a longer-than-average time to reenter the cell cycle after arresting in G0, suggesting that Bcl-xL and Bcl-2 interact with components of the cell-cycle machinery.
3.4. Tumor Suppressor Genes and Apoptosis
Tumor suppressor gene products suppress unrestrained cell proliferation through their specific inhibitory effects on the cell cycle and are implicated in the development of neoplasia following their loss or functional inactivation. One tumor suppressor gene product, p53, induces apoptosis in some tumor cell lines. p53 is a short-lived protein that is stabilized in the presence of DNA damage and triggers cell-cycle arrest, presumably to facilitate DNA repair (62). If the DNA damage is too great to repair, apoptosis is triggered, earning p53 the title “guardian of the genome” (63). At present, it is unclear if p53's induction of apoptosis is effected through its upregulation of p21Waf-1/Cip-1, leading to cell-cycle arrest (64), through some other p53-regulated gene, or is independent of p53 transcriptional activity (65). Bax, the pro-apoptotic Bcl-2 homologue, is transcriptionally regulated by p53, but not all apoptotic cell deaths induced by p53 stabilization require Bax expression (66.( The role of p53 as a sensor of DNA damage was neatly illustrated by examining apoptosis in thymocytes from p53-knockout mice (67, 68). p53 null thymocytes undergo death by apoptosis as normal, except when treated with agents that damage DNA, such as etoposide or irradiation. Therefore, in the absence of p53, the cell is neither instructed to leave the cell cycle and repair the damaged DNA, nor to die. Cells therefore progress through subsequent cycles with damaged and mutated DNA. The role of p53 in inducing apoptosis in circumstances in which DNA damage is not evident is at present unclear, although in p53 knockout mice there is no evidence of a defect in tissue homeostasis due to ineffective cell death when cytokines become limiting (67, 68).
A second tumor suppressor gene implicated in control of apoptosis is the retinoblastoma (rb) gene (69-71). The absence of the functional gene product, Rb, results in massive apoptosis in the haemopoietic and nervous system possibly due to the cells being constantly in cycle and unable to arrest or terminally differentiate. This makes it impossible for cells to establish appropriate survival signals and cell:cell contacts. Loss of both p53 and Rb function results in rapid tumor progression (72), suggesting that cells which have overcome restraints upon cell-cycle progression can also escape apoptosis, underlining the link between cell proliferation and cell death.
3.5. Cytokine Signaling Pathways and Apoptosis
Although a wealth of information is available about the signal transduction pathways activated by cytokines and growth factors in many cell types, surprisingly little is known about how apoptosis ) and conversely cell viability) is modulated. Initially, many of the factors now referred to as survival factors were considered solely to stimulate cells to proliferate. Thus, many of the identified signal transduction pathways are involved in proliferation but not necessarily survival. For survival signals, two pathways can exist. Cells can be stimulated to survive by one specific pathway. Loss of this signal upon removal of the cytokine may simply trigger cell death due to negation of the first signal. However, removal of the cytokine may trigger another independent signaling pathway. Only now are these different possibilities being investigated.
3.5.1. Survival Signals
Cytokines with very different actions activate substantially overlapping signaling circuits, many of which appear to be required, but are not solely responsible, for regulating cell survival. However, even with these complex signaling networks, it is possible to gather some information about survival signals. For example, apoptosis stimulated by the withdrawal of nerve growth factor (NGF) in phaeochromocytoma (PC12) cells involves the induction of the AP-1 transcription factor c-Jun proximal to the time when neurons become committed to apoptosis. Activation of c-Jun requires phosphorylation by the protein kinase JNK (MKK4) that is, in turn, activated by MAP kinase (73) (see Phosphorylation, Protein). In contrast, the suppression of apoptosis by NGF in the same cells may involve a discrete survival signaling pathway routed through Ras (74), Raf, and MAP kinase (73) . Hence, in this case, the induction of apoptosis and promotion of survival may be independent informational processes, rather than mere negation of one other.
In some documented cases, the ability of anti-apoptotic cytokines to suppress apoptosis does not depend on the synthesis of new genes or proteins, indicating that survival is mediated by posttranslational mechanisms. IGF-1 (25), a potent inhibitor of apoptosis in many cell types, and epidermal growth factor (EGF) (75) are reported to suppress apoptosis effectively in cells treated with inhibitors of RNA and protein synthesis. The suppression of apoptosis in such instances must be effected by means of preexisting molecular machinery. One possible candidate signal is the activation of PI-3 kinase (PI3-K) through Ras, whose inhibition blocks the ability of NGF to mitigate apoptosis (76). However, specificity is absent, since PI3-K is activated in many signaling pathways in response to signals that do not suppress apoptosis. IGF-1 also signals via PI3-K, and the pro- and anti-apoptotic signaling pathways for this cytokine have been identified. PI3-K can be activated by its upstream effector Ras, and it can activate several downstream pathways, including the ribosomal protein p70S6K, the Rho family polypeptide Rac, and the serine/threonine protein kinase PKB/Akt. Of these downstream effectors, PKB/Akt is anti-apoptotic in fibroblasts expressing c-Myc and in other cell types in the absence of survival factors, whereas the Ras-mediated Raf signaling pathway is pro-apoptotic (77-79). These findings underscore the pleiotropic nature of intracellular signaling. Signals emanating from GTP-Ras trigger a plethora of potential biological outcomes, some of which promote apoptosis and some of which suppress it. The net outcome for Ras activation is presumably dictated by downstream interactions that potentiate or mitigate other signals.
3.5.2. Killer Cytokines
Apoptosis can also be triggered by certain cytokines, such as TNFa and CD95 ligand. These two pathways represent the best understood triggers of apoptosis, since their activation pathways are essentially mapped. CD95 ligand and TNFa bind and activate their specific receptors on the surface of the cell (80). The TNF type 1 and CD95 (Fas, Apo-1) receptors are functionally similar; both are transmembrane receptors that contain a region within their cytoplasmic tail which is necessary for the initiation of the apoptotic response (81-83). This region of homology is called the death domain ) DD). Several adaptor proteins bind to this domain and can trigger apoptosis by directly linking to the downstream effector caspase machinery (23). Alternatively, in the case of TNFR1, the adaptor proteins can also activate NFkB signaling pathways (84). CD95-induced cell death, eg, results in the association of the death-induced signaling c omplex (DISC) (80). Upon binding of its ligand, the CD95 receptor trimerizes and binds to its death domain the protein Fas-associated death domain (FADD), also known as MORT1 (85, 86). FADD contains a C-terminal DD and, at its N-terminus, a death effector domain (DED), so called because the expression of this domain is required to trigger CD-95-induced apoptosis. The DED domain of FADD binds to the DED in caspase-8, a protein also called FLICE (FADD-like ICE, where ICE is “interleukin converting enzyme”) (87, 88). The thiol proteinase domain of FLICE, located at its C-terminus, is activated upon binding FADD, and FLICE then activates the caspase cascade by cleaving downstream caspases (89). CD95-triggered apoptosis can be suppressed by expression of the anti-apoptotic proteins Bcl-2 and Bcl-xL (90) or by proteins that inhibit the interaction of FLICE with FADD (91). Thus, the binding of CD95 ligand to the receptor and recruitment of FADD occur within the decision phase of apoptosis. The commitment of cells to CD95-induced apoptosis does not appear to occur until FLICE is recruited to the DISC, cleaved, and therefore activated. TNFR1 exhibits a similar pathway involving an overlapping set of adaptor proteins (23).
4. The Commitment Phase of Apoptosis
As yet, it is unclear what commits a cell to undergo apoptosis. To a certain degree, the final trigger must be cell autonomous, because cells that have received the same stimulus do not all enter apoptosis at the same time; cells may die within 10 minutes of the signal or within 3 days. Initially, the caspases were proposed to act at this point, with their activation leading to certain death (92-94( Although exogenous expression of pro-caspases leads to cell death, this death is again stochastic and can be suppressed by the addition of IGF-1 (95) or thiol proteinase inhibitors such as the poxvirus protein CrmA and baculovirus inhibitor p35 (96, 97). Surprisingly though, not all caspase inhibitors suppress cell death. In the case of c-Myc-induced or Bax-induced apoptosis, the addition of tetrapeptide caspase inhibitors based on the pro-IL-1b cleavage site does not prevent cells from becoming committed to die (98, 99). Thus, even when caspases are inhibited, the cell still undergoes commitment, but it takes much longer to progress through the morphological changes of apoptosis (99) .This result suggests caspases are required for the rapid execution phase of apoptosis, but not necessarily for commitment.
The unknown entity in mammalian cells is the putative homologue of the cell death gene 4 (Ced-4( . Ced-4 is required for cell death in C. Elegans and appears to function upstream of Ced-3, suggesting that Ced-4 may be involved in commitment (100). Recent data have shown that Ced-9, Ced-3, and Ced-4 bind one another, with Ced-4 being the critical intermediate (101-104) . Without Ced-4 expression, Ced-9 does not interact with Ced-3. The expression of Ced-4 in mammalian cells produces a similar reaction between Bcl-xL and the Ced-3 homologue, FLICE (caspase-8) (101). Whether Ced-4 is a killer of mammalian cells is also not clear, but Ced-4 expression is toxic to yeast (104). In the fission yeast Schizosaccharomyces pombe, Ced-4 localizes to condensed chromatin and induces caspase-independent death in this apoptotically naïve cell model. Cell death is suppressed upon the expression of Ced-9, which causes relocalization of Ced-4 to the membrane of the nucleus and to the endoplasmic reticulum. These data strongly suggest that Ced-9 suppresses Ced-4-induced death by binding to it. This, in turn, implies that the disruption of the Bcl-xL/ Ced-4-“like protein” and FLICE interaction in mammalian cells may commit the cell to death.
5. The Execution Phase of Apoptosis
The execution phase describes the stage where the cells morphologically change and exhibit all the characteristics of apoptosis. Not all the agents responsible for these changes have been identified, but activation of the endonuclease occurs at this stage, as does activation of the ced-3 homologues, the caspases. Overall, the caspase pathway can be thought of as a packaging process that carries out the rapid dismantling of the cell. Cells still die in the absence of caspase activity, but death is significantly prolonged (99). The cleavage of caspase substrates such as Poly-(ADP ribose( polymerase (PARP), the catalytic subunit of DNA protein kinase, and nuclear lamins [reviewed in (105)], in concert with an endonuclease, allow the nucleus and chromatin to be degraded efficiently. This saves the dying cell energy and facilitates the maintenance of membrane integrity, which in turn prevents release of the cell contents, which would stimulate an immune response.
Other effectors of the execution phase are not so well documented. Both ceramide and reactive oxygen species are found in apoptotic cells, but their exact roles are not clear (110-114): They may well be a consequence of death, rather than an effector. Changes in mitochondria also occur within apoptotic cells (115-117). Selective release of cytochrome c from the mitochondria appears to trigger apoptosis through activation of the caspases. A membrane permeability transition also occurs where the pores that regulate traffic between the mitochondrial outer and inner membranes become disrupted (117). However, it is unclear exactly when this change occurs within an individual cell undergoing apoptosis, making it difficult to assign these changes to a particular phase within the apoptotic pathway.
Overall, the execution phase is one where caspases and endonucleases are active and the cell is dismantled, progressing through the series of morphological changes that originally defined the process of apoptosis. Execution ends with the efficient and rapid phagocytosis of the corpse.
6. Summary
Apoptosis is an active form of cell death indistinguishable from programmed cell death that occurs during invertebrate and vertebrate development. Apoptosis is thought to be the default state of all cells; hence, cells must receive a survival signal in order to remain viable. Apoptosis is triggered in response to a diverse set of stimuli and plays a critical role in the maintenance of tissue homeostasis. Mutations that disrupt the apoptotic pathway are pivotal in the development of many diseases, including degenerative brain disorders and cancer.
References
1. A. H. Wyllie, J. F. Kerr, and A. R. Currie (1972) Cellular events in the adrenal cortex following ACTH deprivation. J. Pathol. 106, 1.
2. C. B. Thompson (1995) Apoptosis in the pathogenesis and treatment of disease. Science 267, 1462-1456 .
3. G. Evan, E. Harrington, A. Fanidi, H. Land, B. Amati, and M. Bennett (1994) Integrated control of cell proliferation and cell death by the c-myc oncogene. Phil. Trans. R Soc. Lond. B 345 , 269–275
4. M. D. Jacobson, M. Weil, and M. C. Raff (1997) Programmed cell death in animal development. Cell 88, 347–354.
5. J. R. Hinchliffe and D. A. Ede (1973) Cell death and the development of limb form and skeletal pattern in normal and wingless (ws) chick embryos. J. Embryol. Exp. Morphol. 30, 772-753 .
6. A. H. Wyllie, J. F. Kerr, and A. R. Currie (1980) Cell death: The significance of apoptosis. Int. Rev. Cytol. 68, 251–306.
7. J. F. Kerr, A. H. Wyllie, and A. R. Currie (1972) Apoptosis: A basic biological phenomenon with wide–ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257.
8. A. H. Wyllie (1980) Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 284, 555–556.
9. J. Savill, V. Fadok, P. Henson, and C. Haslett (1993) Phagocyte recognition of cells undergoing apoptosis. Immunol. Today 14, 131–136. 10. J. Savil, I. Dransfield, N. Hogg, and C. Haslett (1990) Vitronectin receptor-mediated phagocytosis of cells undergoing apoptosis. Nature 343, 170–173.
11. J. Savill, N. Hogg, Y. Ren, and C. Haslett (1992) Thrombospondin cooperates with CD36 and the vitronectin receptor in macrophage recognition of neutrophils undergoing apoptosis. J. Clin. Invest. 90, 1513–1522.
12. Y. Ren, R. L. Silverstein, J. Allen, and J. Savill (1995) CD36 gene transfer confers capacity for phagocytosis of cells undergoing apoptosis. J. Exp. Med. 181, 1857–1862.
13. V. A. Fadok, D. R. Voelker, P. A. Campbell, D. L. Bratton, J. J. Cohen, P. W. Noble, D. W. Riches, and P. M. Henson (1993) The ability to recognize phosphatidylserine on apoptotic cells is an inducible function in murine bone marrow-derived macrophages. Chest 103, 102s.
14. V. A. Fadok, D. R. Voelker, P. A. Campbell J. J. Cohen, D. L. Bratton, and P. M. Henson (1992) Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 148, 2207–2216.
15. V. A. Fadok et al. (1992) Different populations of macrophages use either the vitronectin receptor or the phosphatidylserine receptor to recognize and remove apoptotic cells. J. Immunol. 149, 4029–4035.
16. P. K. Flora and C. D. Gregory (1994) Recognition of apoptotic cells by human macrophages: Inhibition by a monocyte/macrophage-specific monoclonal antibody. Eur. J. Immunol. 24, 2632-2625 .
17. M. F. Luciani and G. Chimini (1996) The ATP binding cassette transporter ABC1 is required for the engulfment of corpses generated by apoptotic cell death. Embo. J. 15, 226–235.
18. N. Platt and S. Gordon (1995) Role of the murine scavenger receptor in the recognition of apoptotic thymocytes by macrophages. J. Cell Biochem. Suppl. 19B, 300.
19. M. Stern, J. Savill, and C. Haslett (1996) Human monocyte-derived macrophage phagocytosis of senescent eosinophils undergoing apoptosis. Mediation by alpha v beta 3/CD36/thrombospondin recognition mechanism and lack of phlogistic response. Am. J. Pathol. 149, 911–921.
20. N. J. McCarthy, C. A. Smith, and G. T. Williams (1992) Apoptosis in the development of the immune system: Growth factors, clonal selection and bcl-2. Canc. Metas. Rev. 11, 157–178.
21. G. I. Evan, L. Brown, M. Whyte and E. Harrington (1995) Apoptosis and the cell-cycle. Curr. Opin. Cell Biol. 7, 825–834.
22. M. Raff, B. Barres, J. Burne, H. Coles, Y. Ishizaki, and M. Jacobson (1993) Programmed cell death and the control of cell survival: Lessons from the nervous system. Science 262, 695–700.
23. S. Nagata (1997) Apoptosis by death factor. Cell 88, 355–365.
24. M. K. Collins, J. Marvel, P. Malde, and A. Lopez-Rivas (1992) Interleukin 3 protects murine bone marrow cells from apoptosis induced by DNA damaging agents. J. Exp. Med. 176, 1043– 1051 .
25. E. Harrington, A. Fanidi, M. Bennett, and G. Evan (1994) Modulation of Myc-induced apoptosis by specific cytokines. Embo. J. 13, 3286–3295.
26. G. T. Williams, C. A. Smith, E. Spooncer, T. M. Dexter, and D. R. Taylor (1990( Haemopoietic colony stimulating factors promote cell survival by suppressing apoptosis. Nature 343, 76–79.
27. R. C. Duke and J. J. Cohen (1986) IL-2 addiction: Withdrawal of growth factor activates a suicide program in dependent T cells. Lymphokine Res. 5, 289–299.
28. E. White, P. Sabbatini, M. Debbas, W. Wold, D. Kusher, and L. Gooding (1992) The 19-kilodalton Adenovirus E1B transforming protein inhibits programmed cell death and prevents cytolysis by tumour necrosis factor . Mol. Cell. Biol. 12, 2570–2580.
29. E. White, R. Cipriani, P. Sabbatini, and A. Denton (1991) Adenovirus E1B 19-kilodalton protein overcomes the cytotoxicity of E1A proteins. J. Virol. 65, 2968–2678.
30. A. Zychlinsky, B. Kenny, R. Menard, M. C. Prevost, I. B. Holland, and P. J. Sansonetti (1994( IpaB mediates macrophage apoptosis induced by Shigella flexneri. Mol. Microbiol 11, 619–627 .
31. K. Thirumalai, K. S. Kim, and A. Zychlinsky (1997) IpaB, a Shigella flexneri invasin, colocalizes with interleukin-1 beta-converting enzyme in the cytoplasm of macrophages. Infect. Immun. 65, 787–793.
32. A. Guichon and A. Zychlinsky (1996) Apoptosis as a trigger of inflammation in Shigella-induced cell death. Biochem. Soc. Trans. 24, 1051–1054.
33. G. Evan et al. (1992) Induction of apoptosis in fibroblasts by c-myc protein. Cell 63, 119–125.
34. M. Zonig and G. I. Evan (1996) Cell cycle: On target with Myc. Curr. Biol. 6, 1553–1556.
35. E. Harrington, A. Fanidi, and G. Evan (1994) Oncogenes and cell death. Curr. Opin. Genet. Dev. 4, 120–129.
36. C. A. Evans, L. P. Owen, A. D. Whetton, and C. Dive (1993) Activation of the Abelson tyrosine kinase activity is associated with suppression of apoptosis in hemopoietic cells. Cancer Res. 53, 1735–1738.
37. J. L. Cleveland, M. Dean, N. Rosenberg, J. Y. Wang, and U. R. Rapp (1989) Tyrosine kinase oncogenes abrogate interleukin-3 dependence of murine myeloid cells through signaling pathways involving c-myc: Conditional regulation of c-myc transcription by temperature-sensitive v-abl. Mol. Cell. Biol. 9, 5685–5695.
38. Y. Tsujimoto, L. R. Finger, J. Yunis, P. C. Nowell, and C. M. Croce (1984) Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science 226, 1097–1099.
39. A. Fanidi, E. Harrington, and G. Evan (1992) Cooperative interaction between c-myc and bcl-2 proto-oncogenes. Nature 359, 554–556.
40. R. Bissonnette, F. Echeverri, A. Mahboubi, and D. Green (1992) Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature 359, 552–554.
41. A. Strasser, A. W. Harris, M. L. Bath, and S. Cory (1990) Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 348, 331–333.
42. T. J. McDonnell and S. J. Korsmeyer (1991) Progression from lymphoid hyperplasia to high-grade malignant lymphoma in mice transgenic for the t(14;18). Nature 349, 254–256.
43. S. J. Korsmeyer, T. J. McDonnell, G. Nunez, D. Hockenbery, and R. Young (1990) Bcl-2: B cell life, death and neoplasia. Curr. Top. Microbiol. Immunol. 166, 203–207.
44. Y. Tsujimoto, J. Cossman, E. Jaffe, and C. M. Croce (1985) Involvement of the bcl-2 gene in human follicular lymphoma. Science 228, 1440–1443.
45. J. C. Reed (1997) Double identity for proteins of the Bcl-2 family. Nature 387, 773–776.
46. M. D. Jacobson (1997) Apoptosis: Bcl-2-related proteins get connected. Curr. Biol. 7, R227–R281 .
47. E. Yang and S. J. Korsmeyer (1996) Molecular thanatopsis: A discourse on the BCL2 family and cell death. Blood 88, 386–401.
48. G. Williams and C. Smith (1993) Molecular regulation of apoptosis—genetic-controls on cell-death. Cell 74, 777–779.
49. Z. Oltvai, C. Milliman, and S. Korsmeyer (1993) Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programed cell death. Cell 74, 609–619.
50. L. Boise et al. (1993) bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74, 597–608.
51.T. Chittenden, E. Harrington, R. O''Connor, G. Evan, and B. Guild (1995) Induction of apoptosis by the Bcl-2 homologue Bak. Nature 374, 733–736.
52. M. Kiefer, M. Brauer, V. C. Powers, J. Wu, S. Umansky, L. Tomei, and P. Barr (1995) Modulation of apoptosis by the widely distributed Bcl-2 homologue Bak. Nature 374, 736–739 .
53. S. Farrow et al. (1995) Cloning of a novel bcl-2 homologue by interaction with adnovirus E1B 19K. Nature 374, 731–733.
54. B. S. Chang, A. J. Minn, S. E. Muchmore, S. W. Fesik, and C. B. Thompson (1997( Identification of a novel regulatory domain in Bcl-xL and Bcl-2. Embo J. 16, 968–977.
55. S. W. Muchmore, M. Sattler, H. Liang, R. P. Meadows, J. E. Harlan, H. S. Yoon, D. Nettesheim, B. S. Chang, C. B. Thompson, S. L. Wong, S. L. Ng, and S. W. Fesik (1996) X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature 381, 341-335.
56. X.-M. Yin, Z. Oltvai, and S. Korsemeyer (1994) Bh1 and bh2 domains of Bcl-2 are required for inhibition of apoptosis and heterodimerization with Bax. Nature 369, 321–323.
57. T. Sato, S. Irie, S. Krajewski, and J. C. Reed (1994) Cloning and sequencing of a cDNA encoding the rat Bcl-2 protein. Gene 140, 291–292.
58. S. Takayama et al. (1995) Cloning and functional-analysis of bag-1—a novel bcl-2-binding. Cell 80, 279–284.
59. E. Yang, J. P. Zha, J. Jockel, L. H. Boise, C. B. Thompson, and S. J. Korsmeyer (1995) Bad, a heterodimeric partner for Bcl-xL and Bcl-2, displaces Bax and promotes cell death. Cell 80, 285-291.
60. J. Zha, H. Harada, E. Yang, J. Jockel, and S. J. Korsmeyer (1996) Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell 87, 619–628 (see comments. (
61. L. O''Reilly, D. Huang, and A. Strasser (1996) The cell death inhibitor Bcl-2 and its homologues influence control of cell cycle entry. Embo. J. 15, 6979–6990.
62. M. B. Kastan, O. Onyekwere, D. Sidransky, B. Vogelstein, and R. W. Craig (1991( Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51, 6304–6311 .
63. D. P. Lane (1992) Cancer. p53, guardian of the genome. Nature 358, 15–16 (news; comment. (
64. W. El-Deiry et al. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 76, 817–825 .
65. C. Caelles, A. Helmberg, and M. Karin (1994) p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature 370, 220–223.
66. C. Yin, C. M. Knudson, S. J. Korsmeyer, and T. Van Dyke (1997) Bax suppresses tumorigenesis and stimulates apoptosis in vivo. Nature 385, 637–640.
67. S. W. Lowe, E. M. Schmitt, S. W. Smith, B. A. Osborne, and T. Jacks (1993) p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362, 847–849.
68. A. R. Clarke et al. (1993) Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 362, 849–852.
69. A. Clarke et al. (1992) Requirement for a functional Rb-1 gene in murine development. Nature 359 , 328-330.
70. E.-H. Lee et al. (1992) Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359, 288–294.
71. T. Jacks, A. Fazeli, E. Schmitt, R. Bronson, M. Goodell, and R. Weinberg (1992) Effects of an Rb mutation in the mouse. Nature 359, 295–300.
72. H. Symonds et al. (1994) p53-dependent apoptosis suppresses tumour growth and progression in vivo. Cell 76, 703–711.
73. Z. Xia, M. Dickens, J. Raingeaud, R. Davis, and M. Greenberg (1995) Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 270, 1326–1331.
74. C. D. Nobes and A. M. Tolkovsky (1995) Neutralizing anti-p21ras Fabs suppress rat sympathetic neuron survival induced by NGF, LIF, CNTF and cAMP. Eur. J. Neurosci. 7, 344-350 .
75. A. Geier, R. Beery, M. Haimsohn, R. Hemi, Z. Malik, and A. Karasik (1994) Epidermal growth factor, phorbol esters, and aurintricarboxylic acid are survival factors for MDA-231 cells exposed to adriamycin. In Vitro Cell Dev. Biol. Anim. 30a, 867–874.
76. R. Yao and G. M. Cooper (1995) Requirement for phosphatidylinositol-3 kinase in the prevention of apoptosis by nerve growth factor. Science 267, 2003–2006.
77. S. Kennedy, A. Wagner, S. Conzen, J. Jordan, A. Bellacosa, P. Tsichlis, and N. Hay (1997( The PI 3-kinase/akt signaling pathway delivers an anti-apoptotic signal. Genes & Devel. 11, 701-713 .
78. H. Dudek et al. (1997) Regulation of neuronal survival by the serine-threonine protein-kinase akt. Science 275, 661–665.
79. A. Kauffmann-Zeh, P. Rodriguez-Viciana, E. Ulrich, C. Gilbert, P. Coffer, and G. Evan (1997( Suppression of c-Myc-induced apoptosis by Ras signalling through PI 3-kinase and PKB. Nature 385, 544–548.
80. P. Krammer, I. Behrmann, P. Daniel, J. Dhein, and K.-M. Debatin (1994) Regulation of apoptosis in the immune sustem. Curr. Opin. Immunol. 6, 279–289.
81. S. Nagata and P. Golstein (1995) The Fas death factor. Science 267, 1449–1456.
82. L. Tartaglia, T. Ayres, G. Wong, and D. Goeddel (1993) A novel domain within the 55 kd TNF receptor signals cell death. Cell 74, 845–853.
83. N. Itoh and S. Nagata (1993) A novel protein domain required for apoptosis. Mutational analysis of human Fas antigen. J. Biol. Chem. 268, 10932–10937.
84. M. Rothe, V. Sarma, V. W. Dixit, and D. V. Goeddel (1995) Traf2-mediated activation of NF-kappa-b by TNF receptor-2 and CD40. Science 269, 1424–1427.
85. M. Boldin, E. Varfolomeev, Z. Pancer, I. Mett, J. Camonis, and D. Wallach (1995) A novel protein that interacts with the death domain of Fas/Apo1 contains a sequence related tothea death domain. J. Biol. Chem. 270, 7795–7798.
86. A. M. Chinnaiyan, K. O''Rourke, M. Tewari, and V. M. Dixit (1995) FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell 81, 505–512.
87. M. Muzio et al. (1996) FLICE, a novel FADD homologous ICE/CED-3–like protease, is recruited to the CD95 (Fas/Apo-1) death-inducing signaling complex. Cell 85, 817–827.
88. M. Boldin, T. Goncharov, Y. Goltsev, and D. Wallach (1996) Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 85, 803–815 .
89. A. Chinnaiyan and V. Dixit (1996) The cell-death machine. Curr. Biol. 6, 555–562.
90. X. Zhang et al. (1996) Up-regulation of Bcl-xL expression protects CD40-activated human B cells from Fas-mediated apoptosis. Cell Immunol 173, 149–154.
91. M. Thome et al. (1997) Viral FLICE inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386, 517–521.
92. E. Alnemri, D. Livingston, D. Nicholson, G. Salvesan, N. Thornberry, W. Wong, and J. Yuan (1996) Human ICE/CED-3 protease nomeclature. Cell 87, 171.
93. Y. Lazebnik, A. Takahashi, G. Poirier, S. H. Kaufmann, and W. Earnshaw (1995( Characterization of the execution phase of apoptosis in vitro using extracts from condemned-phase cells. J. Cell Sci. 19, 41–49.
94. A. Takahashi and W. Earnshaw (1996) Ice-related proteases in apoptosis. Curr. Opin. Gen. & Dev. 6, 50–55.
95. Y. Jung, M. Miura, and J. Yuan (1996) Suppression of interleukin-1 beta-converting enzyme-mediated cell death by insulin-like growth factor. J. Biol. Chem. 271, 5112–5117.
96. V. Gagliardini, P. A. Fernandez, R. K. Lee, H. C. Drexler, R. J. Rotello, M. C. Fishman, and J. Yuan (1994) Prevention of vertebrate neuronal death by the crmA gene. Science 263, 826–828.
97. N. Bump et al. (1995) Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science 269, 1885–1888.
98. J. Xiang, D. Chao, and S. Korsmeyer (1996) Bax-induced cell death may not require interleukin-1ß-converting enzyme-like proteases. Proc. Natl. Acad. Sci. USA 93, 14559–14563 .
99. N. McCarthy, M. Whyte, C. Gilbert, and G. Evan (1997) Inhibition of Ced-3/ICE-related proteases does not prevent cell death induced by oncogenes, DNA damage, or the Bcl-2 homologue Bak. J. Cell. Biol. 136, 215–227.
100. J. Yuan and H. R. Horvitz (1992) The Caenorhabditis elegans cell death gene ced-4 encodes a novel protein and is expressed during the period of extensive programmed cell death. Development 116, 309–320.
101. A. M. Chinnaiyan, K. O''Rourke, B. R. Lane, and V. M. Dixit (1997) Interaction of CED-4 with CED-3 and CED-9: A molecular framework for cell death. Science 275, 1122–1126.
102. M. S. Spector, S. Desnoyers, D. J. Hoeppner, and M. O. Hengartner (1997) Interaction between the C. elegans cell-death regulators CED-9 and CED-4. Nature 385, 653–656.
103. D. Wu, H. D. Wallen, and G. Nunez (1997) Interaction and regulation of subcellular localization of CED-4 by CED-9. Science 275, 1126–1129.
104. C. James, S. Gschmeissner, A. Fraser, and G. Evan (1997) Ced-4 induces chromatin condensation in S. pombe and is inhibited by direct physical association with Ced-9. Curr. Biol. 7, 246–252.
105. M. Whyte (1996) ICE/Ced-3 proteases in apoptosis. Trends Cell Biol. 6, 245–248.
106. A. Fraser, N. McCarthy, and G. I. Evan (1996) Biochemistry of cell-death. Curr. Opin. Neurobiol. 6, 71–80.
107. T. Fernandes Alnemri et al. (1996) In-vitro activation of CPP32 and Mch3 by Mch4, a novel human apoptotic cysteine protease containing 2 FADD-like domains. Proc. Natl. Acad. Sci. USA 93, 7464–7469.
108. Y. A. Lazebnik, A. Takahashi, R. D. Moir, R. D. Goldman, G. G. Poirier, S. H. Kaufmann, and W. C. Earnshaw (1995) Studies of the lamin proteinase reveal multiple parallel biochemical pathways during apoptotic execution. Proc. Natl. Acad. Sci. USA 92, 9042–9046.
109. A. Fraser and G. Evans (1996) A license to kill. Cell 85, 781–784.
110. P. J. Hartfield, G. C. Mayne, and A. W. Murray (1997) Ceramide induces apoptosis in PC12 cells. FEBS Lett. 401, 148–152.
111. S. C. Wright, H. Zheng, and J. Zhong (1996) Tumor cell resistance to apoptosis due to a defect in the activation of sphingomyelinase and the 24 kDa apoptotic protease (AP24). Faseb. J. 10, 325–332 .
112. P. Santana et al. (1996) Acid sphingomyelinase-deficient human lymphoblasts and mice are defective in radiation-induced apoptosis. Cell 86, 189–199.
113. G. J. Pronk, K. Ramer, P. Amiri, and L. T. Williams (1996) Requirement of an ICE-like protease for induction of apoptosis and ceramide generation by REAPER. Science 271, 808–810 .
. 114M. Jacobson and M. Raff (1995) Programmed cell-death and bcl-2 protection in very-low oxygen. Nature 374, 814–816.
115. R. M. Kluck, E. Bossy Wetzel, D. R. Green, and D. D. Newmeyer (1997) The release of cytochrome c from mitochondria: A primary site for Bcl-2 regulation of apoptosis. Science 275, 1132–1136
116. J. Yang, X. Liu, K. Bhalla, C. N. Kim, A. M. Ibrado, J. Cai, T. I. Peng, D. P. Jones, and X. Wang (1997) Prevention of apoptosis by Bcl-2: Release of cytochrome c from mitochondria blocked. Science 275, 1129–1132.
117. G. Kroemer, N. Zamzami, and S. A. Susin (1997) Mitochondrial control of apoptosis. Immunol. Today 18, 44–51.



|
|
|
|
دخلت غرفة فنسيت ماذا تريد من داخلها.. خبير يفسر الحالة
|
|
|
|
|
|
|
ثورة طبية.. ابتكار أصغر جهاز لتنظيم ضربات القلب في العالم
|
|
|
|
|
|
|
قسم الشؤون الفكرية يعزز مكتبته بفهارس المخطوطات التركية
|
|
|