


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 12-5-2016
Date: 8-3-2021
Date: 1-1-2021
|
Recombination-Repair Systems in E. coli
KEY CONCEPTS
- The rec genes of E. coli encode the principal recombination-repair system.
- The recombination-repair system functions when replication leaves a gap in a newly synthesized strand that is opposite a damaged sequence.
- The single strand of another duplex is used to replace the gap.
- The damaged sequence is then removed and resynthesized.
Recombination-repair systems use activities that overlap with those involved in genetic recombination. They are also sometimes called postreplication repair because they function after replication. Such systems are effective in dealing with the defects produced in daughter duplexes by replication of a template that contains damaged bases. An example is illustrated in FIGURE 1.
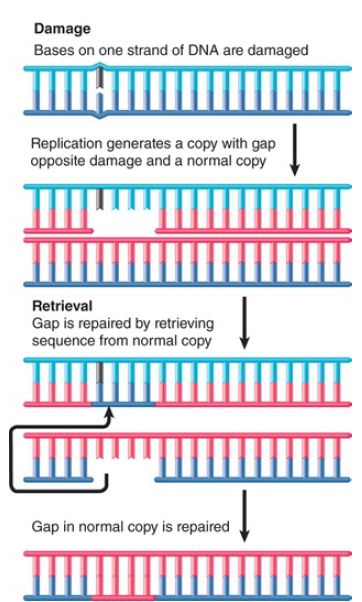
FIGURE 1. An E. coli retrieval system uses a normal strand of DNA to replace the gap left in a newly synthesized strand opposite a site of unrepaired damage.
Consider a structural distortion, such as a pyrimidine dimer, on one strand of a double helix. When the DNA is replicated, the dimer prevents the damaged site from acting as a template. Replication is forced to skip past it.
DNA polymerase probably proceeds up to or close to the pyrimidine dimer. The polymerase then ceases synthesis of the corresponding daughter strand. Replication restarts some distance farther along. This replication may be performed by translesion polymerases, which can replace the main DNA polymerase at such sites of unrepaired damage . A substantial gap is left in the newly synthesized strand.
The resulting daughter duplexes are different in nature. One has the parental strand containing the damaged adduct, which faces a newly synthesized strand with a lengthy gap. The other duplicate has the undamaged parental strand, which has been copied into a normal complementary strand. The retrieval system takes advantage of the normal daughter.
The gap opposite the damaged site in the first duplex is filled by utilizing the homologous single strand of DNA from the normal duplex. Following this single-strand exchange, the recipient duplex has a parental (damaged) strand facing a wild-type strand.
The donor duplex has a normal parental strand facing a gap; the gap can be filled by repair synthesis in the usual way, generating a normal duplex. Thus, the damage is confined to the original distortion (although the same recombination-repair events must be repeated after every replication cycle unless and until the damage is removed by an excision repair system).
The principal recombination-repair pathway in E. coli is identified by the rec genes (see the chapter titled Homologous and Site-Specific Recombination). In E. coli deficient in excision repair, mutation of the recA− gene essentially abolishes all the remaining repair and recovery facilities. Attempts to replicate DNA in uvr recA cells produce fragments of DNA whose size corresponds with the expected distance between thymine dimers. This result implies that the dimers provide a lethal obstacle to replication in the absence of RecA function. It explains why the double mutant cannot tolerate greater than 1 to 2 dimers in its genome (compared with the ability of a wild-type bacterium to handle as many as 50).
One rec pathway involves the recBC genes and is well characterized; the other involves recF and is not so well defined. They fulfill different functions in vivo. The RecBC pathway is involved in restarting stalled replication forks . The RecF pathway is involved in repairing the gaps in a daughter strand that are left after replicating past a pyrimidine dimer.
The RecBC and RecF pathways both function prior to the action of RecA (although in different ways). They lead to the association of RecA with a single-stranded DNA. The ability of RecA to exchange single strands allows it to perform the retrieval step shown in Figure 1. Nuclease and polymerase activities then complete the repair action.
The RecF pathway contains a group of three genes: recF, recO, and recR. The proteins form two types of complexes: RecOR and RecOF. They promote the formation of RecA filaments on singlestranded DNA. One of their functions is to make it possible for the filaments to assemble in spite of the presence of single-strand binding (SSB) protein, which is inhibitory to RecA assembly.
The designations of repair and recombination genes are based on the phenotypes of the mutants, but sometimes a mutation isolated in one set of conditions and named as a uvr gene turns out to have been isolated in another set of conditions as a rec gene. This illustrates the point that the uvr and rec pathways are not independent, because uvr mutants show reduced efficiency in recombination-repair. We must expect to find a network of nuclease, polymerase, and other activities, which constitute repair systems that are partially overlapping (or in which an enzyme usually used to provide some function can be substituted by another from a different pathway).



|
|
|
|
"عادة ليلية" قد تكون المفتاح للوقاية من الخرف
|
|
|
|
|
|
|
ممتص الصدمات: طريقة عمله وأهميته وأبرز علامات تلفه
|
|
|
|
|
|
|
المجمع العلمي للقرآن الكريم يقيم جلسة حوارية لطلبة جامعة الكوفة
|
|
|