

 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية| Exon Sequences Under Negative Selection Are Conserved but Introns Vary |
|
|
|
Read More
Date: 24-5-2021
Date: 25-4-2016
Date: 7-5-2021
|
Exon Sequences Under Negative Selection Are Conserved but Introns Vary
KEY CONCEPTS
-Comparisons of related genes in different species show that the sequences of the corresponding exons are usually conserved but the sequences of the introns are much less similar.
-Introns evolve much more rapidly than exons because of the lack of selective pressure to produce a polypeptide with a useful sequence.
Is a single-copy structural gene completely unique among other genes in its genome? The answer depends on how “completely unique” is defined. Considered as a whole, the gene is unique, but its exons might be related to those of other genes. As a general rule, when two genes are related, the relationship between their exons is closer than the relationship between their introns. In an extreme case, the exons of two genes might encode the same polypeptide sequence, whereas the introns are different. This situation can result from the duplication of a common ancestral gene followed by unique base substitutions in both copies, with substitutions restricted in the exons by the need to encode a functional polypeptide. As we will see in the chapter titled Genome Sequences and Evolution, where we consider the evolution of the genome, exons can be considered basic building blocks that may be assembled in various combinations. It is possible for a gene to have some exons related to those of another gene, with the remaining exons unrelated. Usually, in such cases, the introns are not related at all.
Such homologies between genes can result from duplication and translocation of individual exons. We can plot the homology between two genes in the form of a dot matrix comparison, as in FIGURE 1. A dot is placed in each position that is identical in both genes. The dots form a solid line on the diagonal of the matrix if the two sequences are completely identical. If they are not identical, the line is broken by gaps that lack homology and is displaced laterally or vertically by nucleotide deletions or insertions in one or the other sequence.
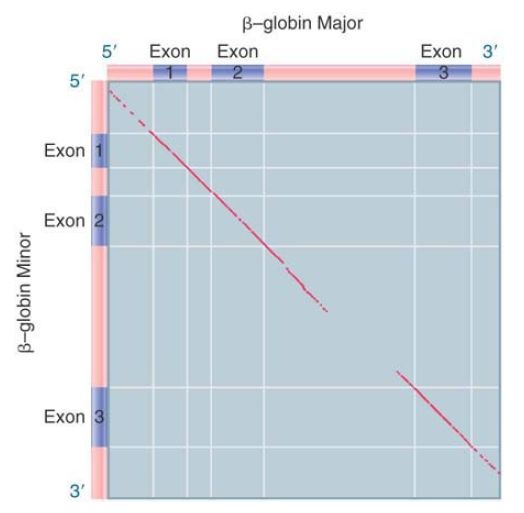
FIGURE 1. The sequences of the mouse bβ - and bβ -globin genes are closely related in coding regions but differ in the flanking UTRs and the long intron.
Data provided by Philip Leder, Harvard Medical School.
When the two mouse β-globin genes are compared in this way, a line of homology extends through the three exons and the small intron. The line disappears in the flanking UTRs and in the large maj min intron. This is a typical pattern in related genes; the coding sequences and areas of introns adjacent to exons retain their similarity, but there is greater divergence in longer introns and in the regions on either side of the coding sequence.
The overall degree of divergence between two homologous exons in related genes corresponds to the differences between the polypeptides. It is mostly a result of base substitutions. In the translated regions, changes in exon sequences are constrained by selection against mutations that alter or destroy the function of the polypeptide. In other words, the exon sequences are conserved by the negative selection of individuals in which the sequences have changed (have not been conserved) to result in a phenotype that is less able to survive and produce fertile progeny. For example, if a mutation in an exon of a gene encoding a crucial enzyme destroys the function of that enzyme, those individuals that carry the mutation (if diploid, then in homozygous form) either do not survive or are otherwise severely affected. The new mutation does not persist.
Many of the preserved changes do not affect codon meanings because they change a codon into another for the same amino acid (i.e., they are synonymous substitutions). In this case, the polypeptide will not change and negative selection will not operate on the phenotype conferred by the polypeptide. Similarly, there are higher rates of change in untranslated regions of the gene (specifically, those that are transcribed to the 5′ UTR [leader] and 3′ UTR [trailer] of the mRNA).
In homologous introns, the pattern of divergence involves both changes in length (due to deletions and insertions) and base substitutions. Introns evolve much more rapidly than exons whenthe exons are under negative sel ection pressure. When a gene is compared among different species, there are instances in which its exons are homologous but its introns have diverged so much that very little homology is retained. Although mutations in certain intron sequences (branch site, splicing junctions, and perhaps other sequences influencing splicing) will be subject to selection, most intron mutations are expected to be selectively neutral.
In general, mutations occur at the same rate in both exons and introns, but exon mutations are eliminated more effectively by selection. However, because of the low level of functional constraints, introns can more freely accumulate point substitutions and other changes. Indeed, it is sometimes possible to locate exons in uncharted sequences by virtue of their conservation relative to introns (see the chapter The Content of the Genome).
From this description it is all too easy to conclude that introns do not have a sequence-specific function. Genes under positive selection, however, cast a different light on the problem.



|
|
|
|
"عادة ليلية" قد تكون المفتاح للوقاية من الخرف
|
|
|
|
|
|
|
ممتص الصدمات: طريقة عمله وأهميته وأبرز علامات تلفه
|
|
|
|
|
|
|
المجمع العلمي للقرآن الكريم يقيم جلسة حوارية لطلبة جامعة الكوفة
|
|
|