النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Ethylene
المؤلف:
D. Grierson, A. J. Hamilton, M. Bouzayen, M. Köck, G. W. Lycett, and S. Barton
المصدر:
In Inducible Plant Proteins
الجزء والصفحة:
5-5-2016
3529
Ethylene
1.History
For a long time, people have recognized the ability of certain gases to stimulate fruit ripening. The ancient Chinese used to burn incense to accelerate ripening. In 1910, Cousins (1) advised the Jamaican Agricultural Department to ship oranges and bananas separately, because some emanation from the oranges (originating from fungal contamination) induced ripening of the bananas. In 1934, Gane (2) proved that ethylene is synthesized by plants and initiates fruit ripening.
The first report of effects of ethylene on vegetative growth dates from 1901, when the Russian physiologist Dimitri Neljubow (3) demonstrated that ethylene in illuminating gas causes the “triple response” in dark-grown pea seedlings: inhibited epicotyl elongation, radial swelling, and a horizontal growth habit, because of loss of geotropism (3). Deleterious effects of illuminating gas had already been observed in the previous century, when leaks in gas pipes caused precocious leaf abscission of surrounding trees.
2. Biosynthesis and Metabolism
Ethylene is a unique gaseous signaling molecule with a simple molecular structure (C2H4). It is active at concentrations as low as 10 nL/L of air (10 ppb). The biological precursor of ethylene in higher plants is L-methionine. By the action of S-adenosyl-L-methionine (AdoMet) synthetase, methionine and ATP are conjugated to form AdoMet (Fig. 1). Subsequently, AdoMet is converted to 1-aminocyclopropane-1-carboxylic acid (ACC) and 5′-methylthio-adenosine (MTA) by the enzyme ACC synthase. MTA is recycled to methionine. ACC is converted to ethylene, HCN, and carbon dioxide by ACC oxidase. Alternatively, ACC can be conjugated to malonyl-ACC and g-L-glutamyl-ACC (4, 5). These inactive derivatives are formed to prevent precursor accumulation and conversion to ethylene. In certain cases, however, malonyl-ACC can also serve as a source of ACC (6).
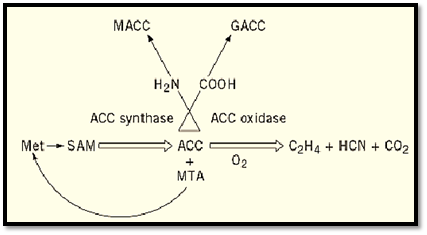
Figure 1. The ethylene biosynthesis pathway in higher plants. ACC, 1-aminocyclopropane-1-carboxylic acid; GACC, glutamyl-ACC; MACC, malonyl-ACC; MTA, 5′-methylthio-adenosine; SAM, S-adenosyl-L-methionine.
Control of ethylene biosynthesis has been observed at the level of ACC synthase and ACC oxidase activities and includes an important aspect of autoregulation (7, 8). In most cases, the ethylene levels are directly correlated with the ACC synthase activity, which is known to be highly regulated during plant development and in response to a complex array of environmental stimuli (9). Data have accumulated, however, that also indicate the importance of ACC oxidase for the regulation of ethylene synthesis (7). The synthesis of ACC does not always translate immediately into ethylene production. Interestingly, ACC is a soluble and translocatable hormone precursor, in contrast to ethylene, which, as a gas, is most probably not suited for targeted translocation processes. Translocation of ACC as a mechanism of interorgan signaling has been proposed for the regulation of plant responses to flooding stress (10, 11).
Because of its low abundance and high lability, ACC synthase proved very difficult to purify. Cloning of ACC synthase genes was achieved independently in several laboratories (12-14). The first nucleotide sequences of ACC synthase cDNA clones were reported by Van Der Straeten et al. (13) and Nakajima et al. (14). The former group obtained peptide sequences after 5000-fold purification of wound-induced pericarp tissue of ripening tomatoes, followed by oligonucleotide-directed screening of a cDNA library (13). Nakajima et al. (14) used an immunological approach to clone ACC synthase from a winter squash cDNA library. Sato and Theologis (12) isolated an ACC synthase clone by screening a cDNA expression library from zucchini with an antibody generated against purified ACC synthase, and the sequence was reported by Sato et al. (15). To date, clones encoding ACC synthase have been isolated from a large array of plant species. The enzyme is encoded by a highly divergent multigene family, and the expression of each gene is differentially regulated by a different subset of inducing or repressing conditions (16). Arabidopsis thaliana was shown to have at least five ACC synthase genes (17, 18). The AT-ACS1 gene expression pattern was analyzed in detail using promoter-b-glucuronidase reporter gene experiments (19). It was shown that AT-ACS1 expression was high in young and immature leaf tissues, lower in stems and in senescing tissues. The transcript levels of most ACC synthase genes are inducible by cycloheximide (16, 17), which suggests either that the messenger RNAs are labile or that transcription of ACC synthase genes is under the control of a labile repressor molecule (17). In either case, it can be concluded that ACC synthase activity is regulated predominantly at the transcriptional level. In addition, the regulation of some ACC synthase genes by various inducers may involve protein kinase activity, which in turn may be controlled through inositol-1,4,5-triphosphate (IP3)-mediated Ca2+ mobilization (20).
A better understanding of the mechanisms involved in the transcriptional regulation of ACC synthase genes will create the possibility to control ethylene formation in a highly specific manner. To date, two strategies have been successfully applied to lower the level of ACC available for ethylene synthesis. In one experiment, the overexpression of the ACC deaminase gene of Pseudomonas sp. strain 6G5 in tomato plants resulted in a reduction of ethylene synthesis (21). Another approach was followed by Oeller et al. (22): expression of a tomato ripening-specific ACC synthase gene in an antisense orientation repressed ethylene synthesis almost completely. These antisense tomato fruits never get ripe until they are treated with ethylene or propylene.
Numerous attempts were made to purify the enzyme downstream of ACC synthase, namely ACC oxidase, or ethylene-forming enzyme. A clone encoding this enzyme was finally isolated in a reverse genetics approach (23). As a first step, ripening-related clones from a tomato cDNA library were isolated. For one of these clones, induction of the corresponding mRNA was tightly correlated with increased ethylene synthesis (24). Transgenic tomato plants expressing the cDNA in an antisense orientation were constructed, resulting in a reduction of ethylene synthesis in a gene dosage-dependent manner. In addition, these plants were delayed in fruit ripening and leaf senescence, and ethylene synthesis was also substantially reduced in wounded leaves (23, 25). Finally, the cDNA was proven to encode ACC oxidase by heterologous expression in yeast and in Xenopus laevis oocytes (26, 27) .
With the exception of preclimacteric fruits and flowers, it was generally believed that ACC oxidase is present constitutively in plant tissues and, therefore, would not contribute to the regulation of ethylene synthesis (28). However, Grierson et al. (7) have pointed out that the rapid induction of ACC oxidase transcripts upon wounding, when assayed in dissected plant organs, may have contributed to the false belief that this enzyme is present constitutively in all tissues. To date, we know that the expression of ACC oxidase genes can be highly regulated throughout plant development (29). As for the expression of ACC synthase genes, the transcriptional regulation of ACC oxidases was also found to be controlled by phosphorylation and dephosphorylation events (30) . Whereas ACC synthase expression was both feedback-inhibited and stimulated by ethylene, (8) ACC oxidase was rapidly induced by ethylene treatments (31). Finally, it should be mentioned that ethylene can be metabolized to ethylene oxide (32).
3. Signal Perception and Transduction
Using the triple response for isolating ethylene mutants, and subsequent double-mutant analysis, the ethylene signal was found to be perceived through several receptors (ETR1, ERS, and maybe also EIN4 and ETR2) and was transduced via a linear pathway to the nucleus, where the expression of a set of primary ethylene-response genes is regulated (33, 34) (Fig. 2). Characterization of the ETR1 gene suggested an explanation for the fact that different ethylene concentrations, which give different responses, can act through a common signal transduction pathway. The ETR1 gene has significant homology with bacterial two-component regulators (35). This kind of receptor systems is known to be able to operate over a wide range of ligand concentrations, resulting in an output that is graded with stimulus intensity (36). The amino-terminal region of the protein contains hydrophobic stretches, each of which is predicted to span the membrane. ETR1 is a membrane-associated, disulfide bond-linked dimer in extracts of Arabidopsis and when expressed in yeast (37). Ethylene was shown to bind to the amino-terminal hydrophobic region (38), and all of the known etr1 mutations were also located in this domain. In addition, the etr1-1 mutation was shown to disrupt ethylene binding. These data are consistent with the chemical properties of ethylene, which is more soluble in lipids than in water and therefore will bind preferentially in a membrane environment (39). The dominant nature of etr1 mutations, together with the redundancy of receptors, suggests that ETR1 inhibits the ethylene response in the absence of ethylene. This characteristic implies that etr1 mutations are gain-of-function mutations, where the enzyme is locked in a catalytically active state. When ethylene binds to the receptor, this inhibitory activity is expected to be disrupted, allowing the ethylene response to occur. Accordingly, mutations preventing the binding of ethylene would then cause a constitutive inhibition of all ethylene responses.
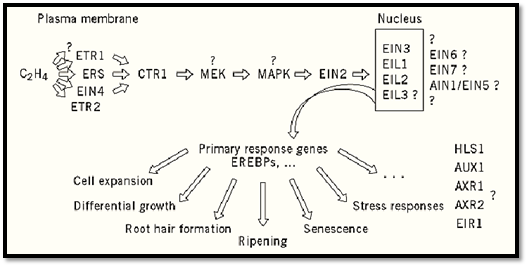
Figure 2. Order of action of components of the ethylene response pathway in Arabidopsis. The pathway was established by double-mutant analysis. EIL3 is homologous to EIN3, EIL1, and EIL2; however, it has not yet been shown to be functional in ethylene signal transduction. The position of AIN1/EIN5, EIN6, and EIN7 in the pathway is unclear. AUX1, AXR1, AXR2, EIR1, and HLS1 control a subset of ethylene responses and may act in parallel to the response pathway defined by the ETR1, CTR1, and EIN2 genes.
Recently, another putative Arabidopsis ethylene receptor, ERS (ethylene response sensor), was cloned (40), supporting the finding that the Arabidopsis genome encodes a complex family of these signal transduction proteins (35). The EIN4 (ethylene-insensitive 4) gene has not yet been isolated. However, as for etr1 mutations, ein4 was dominant, and ein4 ctr1 double mutants had a Ctr1 phenotype, indicating that EIN4 also acts upstream of CTR1 (41). Therefore, it is possible that EIN4 belongs to the same gene family as ETR1 and ERS. It should also be noted that biochemical evidence supports the existence of several ethylene-binding sites that can be classified in two types according to their rate constants for binding (42).
Downstream, the signal is transduced via CTR1, which encodes a Raf-like protein kinase (43). This observation confirms that ethylene induces a transient phosphorylation of several proteins in tobacco (44) .All of the ctr1 mutations found are predicted to disrupt the kinase activity (43). Furthermore, the homology of CTR1 to Raf-1 suggested that a MAP kinase cascade is located downstream (45). A similar combination of a “bacterial” histidine kinase and a Raf-like kinase was found for the high osmolarity glycerol pathway in yeast (46, 47). Signal transduction pathways that include Raf normally contain a MEK protein kinase, which is activated by Raf through phosphorylation (48.( Subsequently, activated MEK phosphorylates a MAP kinase that, in turn, phosphorylates several downstream components. Such a MAP kinase was identified for the yeast high osmolarity glycerol pathway (49) and is also expected in the ethylene response pathway (45). To date, ethylene mutants identifying components of a putative MAP kinase cascade have not been isolated; however, screening for triple-response mutants has not yet saturated the genome (34). Given that ctr1-1 mutations are recessive, it is expected that CTR1 is an inhibitor of the ethylene response in the absence of ethylene. If this were the case, CTR1 may be directly activated by ETR1, which in turn would be active in the absence of ethylene (34).
After CTR1, the ethylene response pathway proceeds with EIN2, followed by EIN3, EIL1, and EIL2. The order of action was based on the fact that ein2 mutations tend to result in strong insensitivity, in contrast to ein3 mutants, which remain more responsive to ethylene even in the case of null alleles (33, 41). This characteristic was confirmed by the cloning of EIN3 and the related genes EIL1, EIL2, and EIL3. Overexpression of EIN3 and EIL1 in an ein2 background resulted in a constitutive ethylene response phenotype (33), which can only be explained when EIN3 and EIL1 act downstream of EIN2, so that mutations in EIN2 do not inhibit the overexpression phenotype. EIN3, EIL1, and EIL2 encode a novel class of nuclear proteins; and they seem to be regulating the same downstream components, because overexpression in each case complemented ein3. These proteins may be directly regulating gene transcription or may interact with transcription factors that have ethylene-regulated genes as targets (33).
A number of weakly insensitive mutants identified genes AIN1/EIN5, EIN6, and EIN7, which act downstream of CTR1, but their position relative to EIN2, EIN3, EIL1, and EIL2 could not be determined (41, 50). In addition, certain mutations affect a specific part of the ethylene response. For example, the HLS1 (HOOKLESS1) gene controls ethylene-regulated differential growth in the apical hook of etiolated seedlings (51), and the EIR1 gene specifically regulates the ethylene sensitivity of roots (41).
The genetic analysis of ethylene signal transduction in Arabidopsis has been very successful and has resulted in the isolation of several components of the pathway. This research will now have to be complemented by biochemical studies and by detailed analysis of the various protein activities in relation to the existing physiological data on ethylene signaling. Additionally, progress is also expected from the use of new screening procedures to isolate novel ethylene mutants (52, 53). The analysis of light-grown populations of mutant seedlings can be complementary to the triple response screening, especially when considering the close interaction between light and ethylene during plant development (39).
4. Downstream Targets
Downstream from EIN3 and EIL1-3, the signal transduction pathway is expected to diverge into several branches that lead to specific ethylene responses. This view is corroborated by the existence of different cis-acting elements that confer ethylene inducibility in response to different inducing conditions. At least two separate mechanisms exist, one of which involves a cis element designated GCC box. In defense-related genes, such as chitinase (54), b-1,3-glucanase (55), and the basic pathogenesis-related (PR1) protein, a common motif was found, with the consensus sequence GCCGCC conserved between the Nicotiana, Arabidopsis, and Phaseolus GCC boxes (56). In addition, a GCC box was found in the promoter of the HSL1 gene (51). Strong evidence for a key role of the GCC box in ethylene-induced transcription was given by coupling a 47-bp fragment from a tobacco b-1,3-glucanase gene containing two GCC box elements to the minimal cauliflower mosaic virus 35 S promoter and by demonstrating that it conveys ethylene responsiveness in transgenic tobacco plants (57). Four different cDNAs encoding GCC box-binding proteins were isolated by southwestern screening of a tobacco cDNA expression library with labeled GCC fragment as a probe (57). The predicted trans factors were named ethylene-response element-binding proteins (EREBPs) and defined a novel class of plant-specific transcriptional regulators. The DNA-binding domain was identified in a region of 59 amino acid residues that was fully conserved in the four EREBPs. Expression of the corresponding genes was ethylene-inducible. Several factors that play a role in development turned out to be members of the EREBP class of transcription factors, including APETALA2 (58), AINTEGUMENTA (59), and TINY (60). More recently, a DNA-binding protein involved in low-temperature expression was shown to be an additional member (61).
A mechanism independent of EREBPs appears to act during senescence of flowers and fruit ripening (62) . A detailed analysis was made of the ethylene-regulated transcription of the glutathione-S-transferase gene 1 (GST1) during carnation petal senescence (63). A cDNA encoding a DNA-binding protein that interacts with a region of GST1 and protects it upon footprinting using deoxyribonuclease 1 was obtained by southwestern screening (64). In tomato ripening, cooperative cis elements are required for ethylene regulation of E4 gene transcription (65).
Finally, it should be noted that all the genes mentioned above are secondary response genes. Primary ethylene-responsive genes, activated within minutes after ethylene induction, remain to be identified.
5. Effects
Ethylene has been shown to participate in a diverse array of plant developmental processes, including seed germination, cell expansion, root initiation, senescence, leaf abscission, and fruit ripening in climacteric fruits (39). In addition, it controls sex determination in some monoecious species. In mango and Bromeliads, it induces flowering. Finally, ethylene is implicated in the control of biotic as well as abiotic stress responses (39).
References
1. H. H. Cousins (1910) III. Agricultural Experiments Citrus. Annual Report of the Department of Agriculture, Jamaica, 7.
2. R. Gane (1934) Nature 134, 1008.
3. D. Neljubow (1901) Beih. Bot. Zentralbl. 10, 128–139.
4. N. Amrhein, D. Schneebeck, H. Skorupka, and S. Tophof (1981) Naturwissenschaften 68, 619–620.
5. M. N. Martin, J. D. Cohen, and R. A. Saftner (1995) Plant Physiol. 109, 917–926.
6. X.-Z. Jiao, S. Philosoph-Hadas, L.-Y. Su, and S. F. Yang (1986) Plant Physiol. 81, 637–641.
7. D. Grierson, A. J. Hamilton, M. Bouzayen, M. Köck, G. W. Lycett, and S. Barton (1992) In Inducible Plant Proteins (J. L. Wray, ed.), Cambridge University Press, Cambridge, U.K., pp. 155-174.
8. R. Fluhr and A. K. Mattoo (1996) Crit. Rev. Plant Sci. 15, 479–523.
9.H. Kende (1989) Plant Physiol. 91, 1–4.
10. K. J. Bradford and S. F. Yang (1980) Plant Physiol. 65, 322–326.
11. M. B. Jackson (1985) Annu. Rev. Plant Physiol. 36, 145–174.
12. T. Sato and A. Theologis (1989) Proc. Natl. Acad. Sci. USA 86, 6621–6625.
13. D. Van Der Straeten, L. Van Wiemeersch, H. M. Goodman, and M. Van Montagu (1990) Proc. Natl. Acad. Sci. USA 87, 4859–4863.
14. N. Nakajima, H. Mori, K. Yamazaki, and H. Imaseki (1990) Plant Cell Physiol. 31, 1021–1029.
15. T. Sato, P. W. Oeller, and A. Theologis (1991) J. Biol. Chem. 266, 3752–3759.
16. T. I. Zarembinski and A. Theologis (1994) Plant Mol. Biol . 26, 1579–1597.
17. X. Liang, S. Abel, J. A. Keller, N. F. Shen, and A. Theologis (1992) Proc. Natl. Acad. Sci. USA 89, 11046-11050.
18. D. Van Der Straeten, R. A. Rodrigues-Pousada, R. Villarroel, S. Hanley, H. M. Goodman, and M. Van Montagu (1992) Proc. Natl. Acad. Sci. USA 89, 9969–9973.
19. R. A. Rodrigues-Pousada, R. De Rycke, A. Dedonder, W. Van Caeneghem, G. Engler, M. Van Montagu, and D. Van Der Straeten (1993) Plant Cell 5, 897–911.
20. X. Liang, N. F. Shen, and A. Theologis (1996) Plant J. 10, 1027–1036.
21. H. J. Klee, M. B. Hayford, K. A. Kretzmer, G. F. Barry, and G. M. Kishore (1991) Plant Cell 3, 1187-1193.
22. P. W. Oeller, L. Min-Wong, L. P. Taylor, D. A. Pike, and A. Theologis (1991) Science 254, 437-439.
23. A. J. Hamilton, G. W. Lycett, and D. Grierson (1990) Nature 346, 284–287.
24. C. J. S. Smith, A. Slater, and D. Grierson (1986) Planta 168, 94–100.
25. S. Picton, S. L. Barton, M. Bouzayen, A. J. Hamilton, and D. Grierson (1993) Plant J. 3, 469–481.
26. A. J. Hamilton, M. Bouzayen, and D. Grierson (1991) Proc. Natl. Acad. Sci. USA 88, 7434–7437.
27. P. Spanu, D. Reinhardt, and T. Boller (1991) EMBO J. 10, 2007–2013.
28. S. F. Yang and N. E. Hoffman (1984) Annu. Rev. Plant Physiol. 35, 155–189.
29. C. S. Barry, B. Blume, M. Bouzayen, W. Cooper, A. J. Hamilton, and D. Grierson (1996) Plant J. 9, 525–535.
30. J. H. Kim, W. T. Kim, B. G. Kang, and S. F. Yang (1997) Plant J. 11, 399–405.
31. W. T. Kim and S. F. Yang (1994) Planta 194, 223–229.
32. I. O. Sanders, A. R. Smith, and M. A. Hall (1986) Physiol. Plant. 66, 723–726.
33. Q. Chao, M. Rothenberg, R. Solano, G. Roman, W. Terzaghi, and J. R. Ecker (1997) Cell 89, 1133-1144.
34. J. J. Kieber (1997) Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 277–296.
35. C. Chang, S. F. Kwok, A. B. Bleecker, and E. M. Meyerowitz (1993) Science 262, 539–544.
36. J. S. Parkinson (1993) Cell 73, 857–871.
37. G. E. Schaller, A. N. Ladd, M. B. Lanahan, J. M. Spanbauer, and A. B. Bleecker (1995) J. Biol. Chem. 270, 12526–12530.
38. G. E. Schaller and A. B. Bleecker (1995) Science 270, 1809–1811.
39. F. B. Abeles, P. W. Morgan, and M. E. Saltveit (1992) Ethylene in Plant Biology, 2nd ed., Academic Press, San Diego, CA.
40. J. Hua, C. Chang, Q. Sun, and E. M. Meyerowitz (1995) Science 269, 1712–1714.
41. G. Roman, B. Lubarsky, J. J. Kieber, M. Rothenberg, and J. R. Ecker (1995) Genetics 139, 1393-1409.
42. N. V. J. Harpham, A. W. Berry, M. G. Holland, I. E. Moshkov, A. R. Smith, and M. A. Hall (1996) Plant Growth Regul . 18, 71–77.
43. J. J. Kieber, M. Rothenberg, G. Roman, K. A. Feldmann, and J. R. Ecker (1993) Cell 72, 427–441.
44. V. Raz and R. Fluhr (1993) Plant Cell 5, 523–530.
45. J. R. Ecker (1995) Science 268, 667–675.
46. I. M. Ota and A. Varshavsky (1993) Science 262, 566–569.
47. T. Maeda, S. M. Wurgler-Murphy, and H. Saito (1994) Nature 369, 242–245.
48. D. R. Alessi, Y. Saito, D. G. Campbell, P. Cohen, G. Sithanandam, U. Rapp, A. Ashworth, C. J. Marshall, and S. Cowley (1994) EMBO J. 13, 1610–1619.
49. F. Posas, S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito (1996) Cell 86,865-875.
50. D. Van Der Straeten, A. Djudzman, W. Van Caeneghem, J. Smalle, and M. Van Montagu (1993) Plant Physiol. 102, 401–408.
51. A. Lehman, R. Black, and J. R. Ecker (1996) Cell 85, 183–194.
52. J. Smalle and D. Van Der Straeten (1997) Physiol. Plant . 100, 593–605.
53. J. Smalle, M. Haegman, J. Kurepa, M. Van Montagu, and D. Van Der Straeten (1997) Proc. Natl. Acad. Sci. USA 94, 2756–2761.
54. K. E. Broglie, P. Biddle, R. Cressman, and R. Broglie (1989) Plant Cell 1, 599–607.
55. G. Felix and F. Meins (1987) Planta 172, 386–392.
56. Y. Eyal, Y. Meller, S. Lev-Yadun, and R. Fluhr (1993) Plant J. 4, 225–234.
57. M. Ohme-Takagi and H. Shinshi (1995) Plant Cell 7, 173–182.
58. D. Weigel (1995) Plant Cell 4, 388–389.
59. K. M. Klucher, H. Chow, L. Reiser, and R. L. Fischer (1996) Plant Cell 8, 137–153.
60. K. Wilson, D. Long, J. Swinburne, and G. Coupland (1996) Plant Cell 8, 659–671.
61. E. J. Stockinger, S. J. Gilmour, and M. F. Thomashow (1997) Proc. Natl. Acad. Sci. USA 94, 1040-1035.
62. J. Deikman (1997) Physiol. Plant. 100, 561–566.
63. H. Itzhaki, J. M. Maxson, and W. R. Woodson (1994) Proc. Natl. Acad. Sci. USA 91, 8925–8929.
64. J. M. Maxon and W. R. Woodson (1996) Plant Mol. Biol. 31, 751–759.
65. R. Xu, S. Goldman, S. Coupe, and J. Deikman (1996) Plant Mol. Biol. 31, 1117–1127.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)