


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 2-5-2016
Date: 18-5-2016
Date: 8-12-2015
|
DNA Repair
DNA repair is the set of enzymatic reactions that correct two types of structural anomalies in DNA (1) .The first class of anomalies involves normal bases in an abnormal sequence context and includes mismatches, loops (extra nucleotides in one strand relative to the complementary strand), and bulges ) a stretch of >1 noncomplementary base within the duplex). The second class, which is referred to as DNA damage or DNA lesions, encompasses abnormal nucleotides (modified, fragmented, cross-linked) within a normal or noncomplementary sequence context.
1. Mismatch Repair
Mismatches and related anomalous structures arise from errors in DNA replication and from recombination between partially homologous (homeologous) sequences. In addition, deamination of methylcytosine spontaneously or by acidic pH is a major cause of converting a G-C base pair to a G · T mismatch (2). The other source of simple mismatches is incorporation errors by DNA polymerases and the presence of a small fraction of bases in the template or in the nucleotide pool in tautomeric forms that lead to abnormal, yet complementary, hydrogen bonding and misincorporation. Loops often result from slippage/dislocation of template or nascent strand at monotonous sequences, such as oligo dA · dT tracts or microsatellite DNA consisting of di- or trinucleotide repeats. Bulges are often caused by recombination between two related sequences with a noncomplementary sequence surrounded by homologous sequences. Mismatches are repaired by two general mechanisms: base-excision repair and the general mismatch repair system (3). In the base-excision repair pathway, the mismatched base (eg, the T in the G · T mismatch) is released by a glycosylase enzyme, and the resulting abasic sugar is removed by the combined actions of AP lyase, which cuts the polynucleotide chain 3′ to the apurinic/apyrimidinic (AP) site, and AP endonuclease, which cuts 5′ to the AP site. The resulting one-nucleotide gap is filled in by a DNA polymerase and sealed by a ligase (see Base Excision Repair). In the general mismatch repair system, an endonuclease incises the newly synthesized strand anywhere from 100 to 1000 nucleotides away and either 5′ or 3′ to the mismatch. The intervening region is removed past the mismatch by exonucleases, and the resulting gap is filled in by DNA polymerase and is ligated to restore the normal duplex.
Defects in the general mismatch repair system (nucleotide excision/long-patch repair) cause hereditary nonpolyposis colorectal cancer (HNPCC) in humans. In addition, mismatch repair defects have been found in several sporadic cancers (3).
2. Damage Repair
DNA damage in the form of base modification, base adduction, base fragmentation, and phosphodiester bond cleavage is caused by many agents including radiation in the form of X-rays (4) ultraviolet (5, 6), and microwave; and chemicals (7) ranging in reactivity from the relatively inert water to highly reactive oxygen radicals and activated polyaromatic hydrocarbons (Table 1). The most common lesions induced in DNA by ionizing radiation and active oxygen species are oxidized and fragmented purines, pyrimidine hydrates and glycols, abasic sites, and single- and double-strand breaks (8, 9). The major lesions induced by ultraviolet light are cyclobutane pyrimidine dimers and [6–4] dipyrimidine photoproducts. There is a virtually infinite variety of lesions produced in DNA by chemical agents. Among those, benzo[a]pyrene-, aflatoxin-, and acetylaminofluorene–guanine adducts are carcinogenic DNA lesions produced by natural and synthetic compounds. Similarly, some anticancer drugs cause DNA damage. Mitomycin C, cisplatin, and cyclophosphamide form monoadducts with guanine bases or cause interstrand cross-links between guanines in each of the two strands of the duplex. Psoralen makes intrastrand thymine monoadducts or interstrand thymine–psoralen–thymine cross-links. Alkylating agents such as methyl methane sulfonate, nitrosoguanidine, and nitrogen– and sulfur–mustard attack essentially all nucleophilic groups in DNA, producing at least 12 different base alkylation adducts and phosphorothioesters (7).
Table 1. Causes and Types of DNA Lesions and Mechanisms of DNA Repair

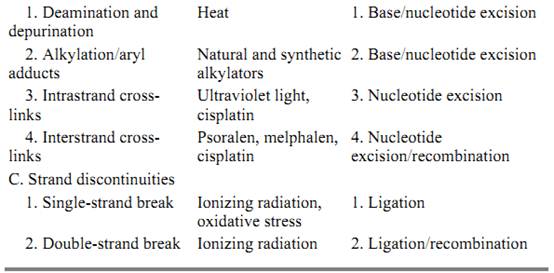
DNA lesions in the form of modified or adducted bases, interstrand cross-links, or discontinuity in one (nick) or both (break) strands interfere with replication, transcription and recombination of DNA; they also cause mutations or cell death, due to the inability to replicate or to a mutation in an essential gene. Furthermore, because many of the agents that cause damage are present in the cell's normal habitat, DNA damage is a frequent occurrence that the cell must deal with on a continuous basis in order to maintain its integrity and survival (9). Hence, cells possess many biochemical reaction pathways (DNA repair mechanisms) to eliminate damage from their chromosomes. These pathways fall into three main categories (1, 10): (i) base excision, (ii) nucleotide excision (or simply excision), and (iii) direct repair mechanisms. As should be apparent from this enumeration, there are extensive functional similarities between the mismatch and damage repair systems. This is not surprising, because the double-helical nature of DNA dictates the types of operations that can be applied to it to remove a mismatched or a damaged base: The mismatch or damage may be removed in the form of a base (11, 12) or a (oligo)nucleotide (10), which creates a single-stranded gap that can be filled using the intact (correct) strand as a template. However, damage repair includes additional mechanisms in its repertoire. One of these is direct repair (1). In this type of repair the chemical bonds that comprise the lesion are broken to restore the normal bases. An example of this type of repair includes photoreactivation by photolyase, which breaks the two sigma bonds between adjacent thymines to restore the dimer to a thymine dinucleotide (13), and O6-methylguanine DNA methyltransferase, which breaks the single bond between the O6 of guanine and the alkyl group and thus restores the normal base (14( . Ligation of a single-strand or double-strand break that is not accompanied by base loss may also be considered an example of direct repair. Finally, recombinational repair is also a repair mechanism unique to damaged DNA. This repair mechanism is employed by the cell when both strands of the duplex are damaged so that the duplex cannot be fixed by direct repair or by base and nucleotide excision repair systems, because these systems rely on an intact strand in the duplex (1). In this rather elaborate repair mechanism, the damaged region of the duplex is replaced by using a homologous duplex to retrieve genetic material and/or information. This repair system uses the enzymes of both nucleotide excision repair and genetic recombination systems.
References
1. A. Sancar and G. B. Sancar (1988) DNA repair enzymes. Annu. Rev. Biochem. 57, 29–67.
2. T. A. Kunkel (1992) DNA replication fidelity. J. Biol. Chem. 267, 18251–18254.
3. P. Modrich and R. Lahue (1996) Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65, 101–133.
4. M. Dizdaroglu (1992) Measurement of radiation induced damage to DNA at the molecular level. Int. J. Radiat. Biol. 61, 175–183.
5. D. E. Brash (1988) UV mutagenic photoproducts in E. coli and human cells: a molecular genetics perspective on human skin cancer. Photochem. Photobiol. 48, 59–66.
6. S. Tornaletti and G. P. Pfeiffer (1996) UV damage and repair mechanisms in mammalian cells. Bioessays 18, 221–228.
7. B. Singer and J. T. Kusmierek (1982) Chemical mutagenesis. Annu. Rev. Biochem. 51, 655–693.
8. E. S. Henle and S. Linn (1997) Formation, prevention and repair of DNA damage by iron/hydrogen peroxide. J. Biol. Chem. 272, 19095–19098.
9. K. B. Beckman and B. A. Ames (1997) Oxidative decay of DNA. J. Biol. Chem. 272, 19633–19636.
10. A. Sancar (1994) Mechanisms of DNA excision repair. Science 266, 1954–1956.
11. E. Seeberg, L. Eide, and M. Bjoras (1995) The base ecision repair pathway. Trends Biochem. Sci. 20, 391–397.
12. M. L. Dodson, M. L. Michaels, and R. S. Lloyd (1994) Unified catlytic mechanism for DNA glycosylases. J. Biol. Chem. 269, 32709–32712.
13. A. Sancar (1996) No “End of History” for photolyases. Science 272, 48–49.
14. L. Samson (1992) The suicidal DNA repair methyltransferases of microbes. Mol. Microbiol. 6, 825–831.



|
|
|
|
للعاملين في الليل.. حيلة صحية تجنبكم خطر هذا النوع من العمل
|
|
|
|
|
|
|
"ناسا" تحتفي برائد الفضاء السوفياتي يوري غاغارين
|
|
|
|
|
|
|
نحو شراكة وطنية متكاملة.. الأمين العام للعتبة الحسينية يبحث مع وكيل وزارة الخارجية آفاق التعاون المؤسسي
|
|
|