


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 29-10-2015
Date: 16-6-2021
Date: 16-5-2016
|
Diphtheria Toxin
Diphtheria was the first infectious disease shown to be caused solely by a toxin (1). Diphtheria toxin ) DT) is encoded by a gene carried by temperate bacteriophages, such as the o-corynephage, that lysogenize Corynebacterium diphtheriae and make it toxigenic. The toxin gene is activated by the release of an iron-binding repressor protein, which binds to DNA in its holo form and in iron-depleted media is released as the apo form (1). The DT gene includes a 25-residue long amino-terminal leader sequence responsible for the export of DT from the cell (2, 3). After proteolytic removal of the leader sequence, DT is released by C. diphtheriae as a single polypeptide chain of 535 amino acid residues. This chain is rapidly cleaved in vivo by furin (4) and in vitro by different proteinases, within a loop containing three Arg residues that connects the first and second domains ) Fig. 1). After such proteolytic nicking, DT is composed of two-chains: A (193 residues), responsible for the intracellular catalytic activity, and B (342 residues), responsible for binding and membrane translocation. A and B are linked by noncovalent forces and a single interchain disulfide bond. Figure 1 shows the three-domain structural organization of DT (5). The amino terminal domain ( residues 1 to 193) corresponds to the A subunit, which is an ADP-ribosyltransferase with a cleft-shaped NAD-binding cavity harboring the Glu148, His21, Trp50, and Tyr65 residues that are essential for activity (5). NAD binds in such a way that its nicotinamide moiety overlaps the phenolic ring of Tyr65, and its adenine group lies over the indole ring of Trp50.
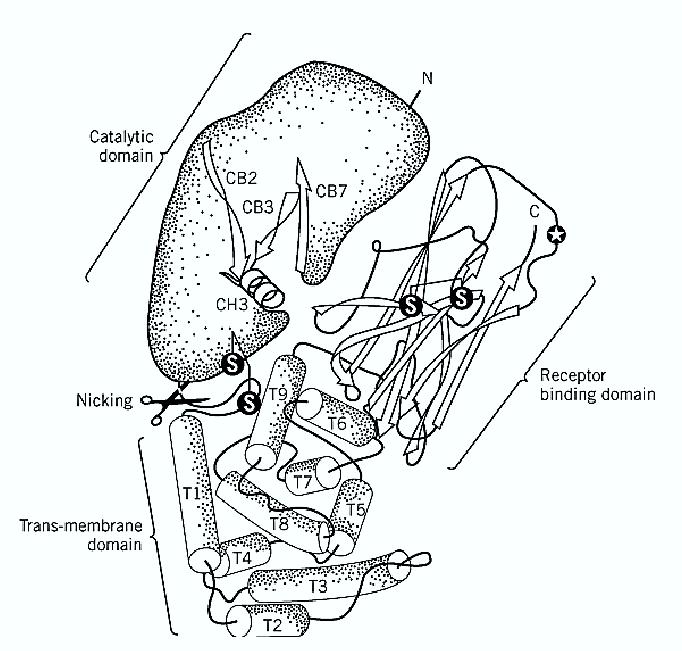
Figure 1. Structure of diphtheria toxin. Diphtheria toxin consists of three domains, characterized by different secondary structure elements and different biological properties. The cleft in the catalytic domain is the NAD binding site. The circled star indicates Ser-508 involved in receptor binding. The toxin is activated by selective proteolysis of a furin-sensitive loop between the A domain and T domain, which is responsible for membrane translocation.
In the nicked toxin, the A domain is disulfide-linked to the T domain (residues 194 to 385), which is involved in the membrane insertion and translocation of DT. The T domain is almost entirely composed of alpha-helices, a type of secondary structure found in the membrane-embedded sector of many integral membrane proteins. The third domain R (residues 386 to 535) is a flattened ß-barrel and responsible for DT binding to its specific receptor, which is a protein that binds heparinlike growth factors (6). The role of this receptor in cell physiology is not known, but it was shown that CD9 enhances the sensitivity of cells to DT (7). The number of DT receptors in different cell lines correlates with their DT sensitivity. Binding is followed by internalization inside clathrin -coated vesicles, which uncoat and merge into early endosomes, a process that requires less than 5 min in Vero cells at 37°C (8). The acidic pH of the endosomal lumen induces a conformational change of DT, with insertion of helices of the T domain into the lipid bilayer (9, 10), to create a transmembrane channel, open laterally to lipids, that mediates the translocation of the A chain into the cytosol (11). It is not known if domain A has to unfold completely in order to pass through the B channel, but it is noteworthy that A can be boiled in SDS and, upon detergent removal, refolds into its active conformation. The translocation and release of A are linked to the reduction of the interchain disulfide bond, which is carried out by yet unidentified reductases. Such reduction is the rate-limiting step of the entire intoxication process (8). Only about a third of the internalized DT molecules are reduced and release their A chain into the cytosol. The remaining, unreduced two-thirds, as well as the B subunits left over on the membrane after A release, are conveyed to the lysosomes and degraded (8).
The A chain of DT ADP-ribosylates, specifically elongation factor 2 (EF-2), which becomes unable to transfer the tRNA-bound amino acid to the growing polypeptide chain, and cell protein biosynthesis are blocked (12). DT-A is active only on the EF-2 of eukaryotes and archaebacteria because they contain diphthamide, a post-translationally modified His residue, which is the unique substrate of DT and of the related exotoxin A released by Pseudomonas aeruginosa (12). The physiological role of diphthamide in vivo is unknown, particularly because cell lines mutated at this His residue, or in one of the enzymes involved in its transformation into diphthamide, appear to grow normally (12). One single molecule of DT is able to kill a cell rather rapidly (13) because it can catalyze the ADP-ribosylation of about 2000 EF-2 per hour, whereas cells in culture produce only about 1500 new EF-2 per hour, and EF-2 synthesis decreases as toxin action progresses. As a result, the lethal dose of DT in sensitive animal species (those possessing DT receptors) is below 0.1 µg DT/kg.
References
1. A. M. Pappenheimer (1982) Harvey Lect. 76, 45–73.
2. L. Greenfield et al. (1983) Proc. Natl. Acad. Sci. USA 80, 6853–6857.
3. G. Ratti, R. Rappuoli, and G. Giannini (1983) Nucleic Acids Res. 11, 6589–6595.
4. J. R. Murphy (1997) In Guidebook to Protein Toxins and Their Use in Cell Biology (R. Rappuoli and C. Montecucco, eds.), Sambrook and Tooze, Oxford University Press, Oxford, UK.
5. S. Choe et al. (1992) Nature 367, 216–222.
6. J. G. Naglich, J. E. Metherall, D. W. Russell, and L. Eidels (1992) Cell 69, 1051–1061.
7. T. Mitamura et al. (1992) J. Cell Biol. 118, 1389–1399.
8. E. Papini, R. Rappuoli, M. Murgia, and C. Montecucco (1993) J. Biol. Chem. 268, 1567–1574.
9. K. Sandvig and S. Olsnes (1981) J. Biol. Chem. 256, 9068–9076.
10. K. J. Oh et al. (1996) Science 273, 810–812.
11. G. Menestrina, G. Schiavo, and C. Montecucco (1994) Mol. Aspects Med. 15, 81–193.
12. R. J. Collier (1990) In ADP-ribosylating Toxins and G Proteins (J. Moss and M. Vaugham,
eds.), American Society for Microbiology, Washington, DC, pp. 3–19.
13. M. Yamaizumi, E. Mekada, T. Uchida, and Y. Okada (1978) Cell 15, 245–250.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
اتحاد كليات الطب الملكية البريطانية يشيد بالمستوى العلمي لطلبة جامعة العميد وبيئتها التعليمية
|
|
|