النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Plate count method ISO 11290-2:1998 Amendment 1:2004 for Listeria monocytogenes in foods
المؤلف:
SILVA, N.D .; TANIWAKI, M.H. ; JUNQUEIRA, V.C.A.; SILVEIRA, N.F.A. , NASCIMENTO , M.D.D. and GOMES ,R.A.R
المصدر:
MICROBIOLOGICAL EXAMINATION METHODS OF FOOD AND WATE A Laboratory Manual
الجزء والصفحة:
15-3-2016
6558
Plate count method ISO 11290-2:1998 Amendment 1:2004 for Listeria monocytogenes in foods
This method of the International Organization for Standardization is applicable to products intended for human consumption or for the feeding of animals. In general the lower limit of enumeration of this method is 10 L. monocytogenes per milliliter of sample for liquid products, or 100 L. monocytogenes per gram of sample for other products.
1. Material required for analysis
Preparation of the sample and serial dilutions
• Diluent: Buffered Peptone Water (BPW) or Half Fraser Broth Base
• Dilution tubes containing 9 ml Buffered Peptone Water (BPW)
Plate count
• Agar Listeria Ottaviani & Agosti (ALOA)
• Glass or plastic spreaders
• Laboratory incubator set to 20 ± 2°C
• Laboratory incubator set to 37 ± 1°C
Confirmation
• Trypticase Soy Agar with 0.6% Yeast Extract
(TSA-YE)
• Trypticase Soy Broth with 0.6% Yeast Extract
(TSB-YE)
• Motility Test Medium ISO (optional)
• 3% Hydrogen Peroxide (for catalase test)
• Gram Stain Reagents
• Sheep Blood Agar plates
• Purple Broth with 0.5% Xylose tubes
• Purple Broth with 0.5% Rhamnose tubes
• L. monocytogenes culture (e.g. NCTC 11994)
• β-hemolytic Staphylococcus aureus culture (e.g. ATCC 25923 or NCTC 1803) (optional)
• Rhodococcus equii culture (e.g. ATCC 6939 or NCTC 1621) (optional)
• Laboratory incubator set to 25 ± 1°C
• Laboratory incubator set to 37 ± 1°C
2. Procedure
A general flowchart for enumeration of L. monocytogenes in foods using the plate count method ISO 11290-2:1998/Amd.1:2004 is shown in Figure 1.
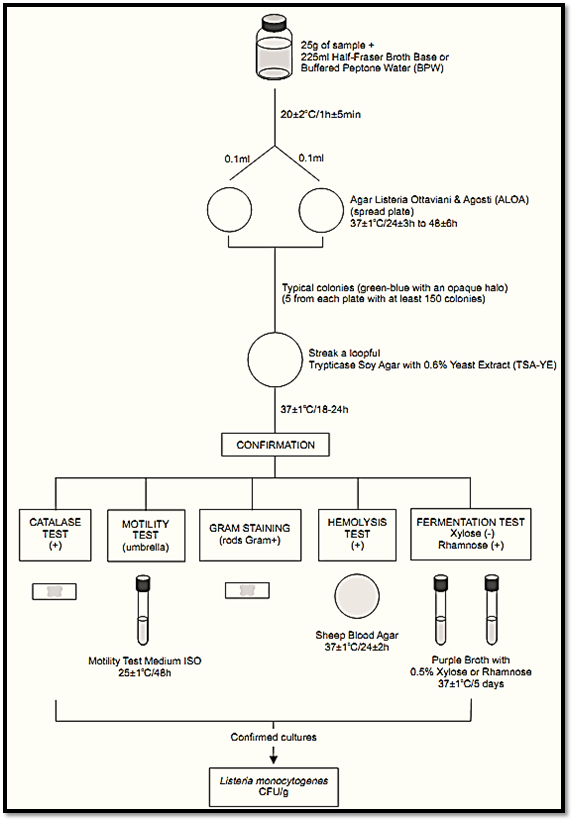
Figure 1 Scheme of analysis for enumeration of L. monocytogenes in foods using the plate count method ISO 11290-2:1998/Amd.1:2004.
Safety precautions recommended by ISO 11290-2:1998/Amd.1:2004: The tests should be performed in properly equipped laboratories, under the supervision of a skilled microbiologist. Great care should be taken in the disposal of contaminated materials. Female laboratory staff should be made aware of the risk to pregnant women. National legislation may involve more specific demands.
a) Preparation of the samples and repair of the injured cells: homogenize 25 g or 25 ml of sample with 225 ml of Buffered Peptone Water (BPW) or Half Fraser Broth Base. Incubate at 20 ± 2°C/1 h ± 5 min (repair of injured cells).
b) Inoculation and incubation: From the BPW (or Half Fraser Broth Base) spread two 0.1 ml aliquots onto two plates of Agar Listeria Ottaviani & Agosti (ALOA). Incubate the plates at 37 ± 1°C/24 ± 3 h-48 ± 6 h.
Note b.1) If the expected count is low, inoculate two 1 ml aliquots of BPW or Half Fraser Broth Base, distributing each 1 ml aliquot onto the surface of three Petri dish or using one large Petri dish (140 mm). If the expected count is high, inoculate decimal dilutions of BPW or Half Fraser Broth Base. If two or more dilutions are inoculated it is not necessary two plates per dilution (ISO 7218:2007).
c) Presumptive count and colony selection for confirmation: Count the typical colonies after 24 ± 3 h of incubation: green-blue surrounded by an opaque halo. If no suspect colonies are evident or if the growth is poor after 24 ± 3 h, re-incubate the plates for an additional 24 h ± 3 h and read after 48 ± 6 h. Select for count the plates contain-ing less than 150 colonies (typical or atypical).
Note c.1) When stressed some strains of L. monocytogenes may show a weak halo or even no halo on ALOA.
Note c.2) The method will not detect strains of L. monocytogenes with slow PIPLC (phosphatidyl inositol phospholipase C) activity (more than four days of incubation are required for these strains).
Select for confirmation five typical colonies from each plate containing less than 150 colonies. If there are fewer than five presumptive colonies on a plate, select all. Purify each culture by streak-ing on a plate of Trypticase Soy Agar with 0.6% Yeast Extract (TSA-YE). Incubate the TSA-YE plates at 37 ± 1°C/18–24 h or until growth is satisfactory.
Select one typical well isolated colony from each TSA-YE plate for the confirmatory tests. Typical colonies on TSA exhibit a bluish color and a granular surface when observed with white light at 45° angle. If the suspect colonies on TSA-YE are not isolated, purify by streaking on a fresh TSA-YE plate before starting the tests.
d) Confirmation: The confirmatory tests are basically the same described in the method BAM/FDA 2011, but there are some variations in the incubation temperature and incubation time:
d.1) Catalase test
d2) Gram stain
d.3) Motility test (optional): From the culture on TSA-YE inoculate a tube of Trypticase Soy Broth with 0.6% Yeast Extract (TSB-YE). Incubate the tube at 25 ± 1°C/8–24 h, prepare a wet-mount and observe microscopically under oil immersion (phase contrast microscopy recommended). Listeria spp. appear as slim, short rods with tumbling motility. Cocci, large rods, or rods with rapid swimming motility are not Listeria spp.
To observe motility using semisolid medium, inoculate the TSA-YE culture into tubes of Motility Test Medium ISO, by stabbing. Incubate the tubes at 25 ± 1°C/48 h and examine for typical umbrella-like growth. If growth is poor after 48 h of incubation, re-incubate the tubes until a total time of five days and re-examine.
Note d.3) According to ISO 11290-2:1998/Amd.1:2004 the motility test is not necessary if the analyst is skilled and regularly works on the detection of L. monocytogenes.
d.4) Hemolysis test: Follow the same procedure described in the method BAM FDA 2011, but the Sheep Blood Agar plates are incubated at 37 ± 1°C/24 ± 2 h. According to ISO 11290-1:1996/Amd.1:2004, is easier to observe the zone of haemolysis if the colony is removed. The hemolysis test may be substituted by the CAMP Test.
d.5) Xylose and rhamnose fermentation test: Follow the same procedure described in the method BAM FDA 2011, but the tubes are incubated at 37 ± 1°C for up to five days, although the positive reactions occur mostly within 24 h to 48 h.
d.6) CAMP Test (Christie-Atkins-Munch-Peter-son Test) (optional): Follow the same procedure described in the method BAM FDA 2011, but the Sheep Blood Agar plates are incubated at 37 ± 1°C/18–24 h. The CAMP test is not required if the hemolysis test results are conclusive.
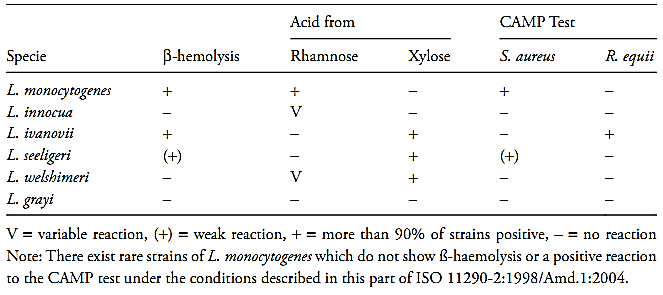
e) Interpretation of the results: Consider as Listeria spp. the small, Gram-positive rods that demonstrate tumbling motility in wet mounts and umbrella-like motility in semisolid medium, generally cata-lase positive. L. monocytogenes are distinguished from other species by the characteristics listed in Table 1.
Table 1 Guide for the interpretation of Listeria monocytogenes confirmatory tests according the method ISO 11290-2:1998/Amd.1:2004
f ) Calculation of results
f.1) First it is necessary to calculate the number of colonies of L. monocytogenes present in each plate retained for count (all the plates containing less than 150 colonies). Use the formula below for this calculation and round the result to a whole number.
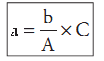
C = total number of typical colonies counted
A = number of typical colonies selected for confirmation
b = number of typical colonies confirmed
a = number of Listeria monocytogenes colonies in the plate.
Example: Total number of typical colonies in a plate (C) = 139; number of typical colonies selected for confirmation (A) = 5; number of colonies con-firmed (b) = 4. Number of L. monocytogenes colonies in the plate (a) = 139 × 4/5 = 111.
f.2) After that it is necessary to calculate the number of L. monocytogenes colony forming units (CFU) per gram or milliliter of the sample, witch is the sum of L. monocytogenes colonies (a) obtained in each plate divided for the sum of sample quantity inoculated in each plate. Use the formula below for this calculation and round the result to a whole number (example 1):

∑ = Sum of L. monocytogenes colonies in
each plate (a1 + a2 + ...)
V = Volume of inoculum in each plate
n1 = number of plate retained at the first dilution counted
n2 = number of plate retained at the second dilution counted
d = dilution factor corresponding to the first dilution retained
If the two dishes, at the level of the initial suspension, contain less than 15 colonies of L. monocytogenes, calculate the result as described above and report the count as estimated (example 2).
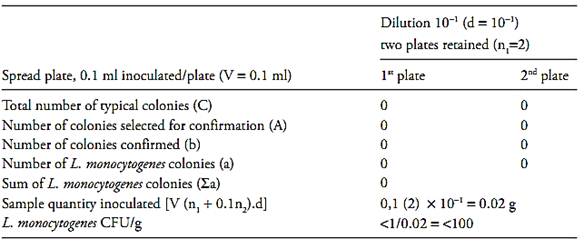
If the two dishes at the level of the initial suspension do not contain any colonies, calculate the result as described above for one colony and report the count as “less than the value obtained” (example 3).
Example 1

Example 2
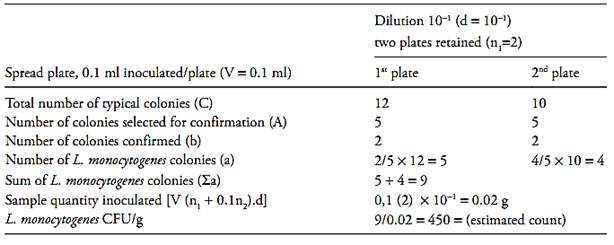
Example 3
References
Silva, N.D .; Taniwaki, M.H. ; Junqueira, V.C.A.; Silveira, N.F.A. , Nasdcimento , M.D.D. and Gomes ,R.A.R .(2013) . Microbiological examination methods of food and water a laboratory Manual. Institute of Food Technology – ITAL, Campinas, SP, Brazil .
International Organization for Standardization (2004) ISO 11290-2:1998/Amd.1:2004. Microbiology of food and animal feeding stuffs– Horizontal method for the detection and enumeration of Listeria monocytogenes – Part 2: Enumeration method. 1st edition:1998, Amendment 1:2004.Geneva, ISO.
International Organization for Standardization (2004) ISO 11290-1:1996/Amd.1:2004. Microbiology of food and animal feeding stuffs – Horizontal method for the detection and enumeration of Listeria monocytogenes – Part 1: detection method. 1st edition:1996, Amendment 1:2004. Geneva, ISO.
 الاكثر قراءة في البكتيريا
الاكثر قراءة في البكتيريا
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)