النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Regulation of Eukaryotic Gene Expression: Coordinate regulation
المؤلف:
Denise R. Ferrier
المصدر:
Lippincott Illustrated Reviews: Biochemistry
الجزء والصفحة:
31-12-2021
2150
Regulation of Eukaryotic Gene Expression: Coordinate regulation
The higher degree of complexity of eukaryotic genomes, as well as the presence of a nuclear membrane, necessitates a wider range of regulatory processes. As with the prokaryotes, transcription is the primary site of regulation. Again, the theme of trans-acting factors binding to cis-acting elements is seen. Operons, however, are not found in eukaryotes, which must use alternate strategies to solve the problem of how to coordinately regulate all the genes required for a specific response.
In eukaryotes, gene expression is also regulated at multiple levels other than transcription. For example, the major modes of posttranscriptional regulation at the mRNA level are alternative mRNA splicing and polyadenylation, control of mRNA stability, and control of translational efficiency. Additional regulation at the protein level occurs by mechanisms that modulate stability, processing, or targeting of the protein.
Coordinate regulation
The need to coordinately regulate a group of genes to cause a particular response is of key importance in organisms with more than one chromosome. An underlying theme occurs repeatedly: A trans-acting protein functions as a specific transcription factor (STF) that binds to a cisacting regulatory consensus sequence on each of the genes in the group even if they are on different chromosomes. [Note: The STF has a DNA-binding domain (DBD) and a transcription activation domain (TAD).
The TAD recruits coactivators, such as histone acetyltransferases , and the general transcription factors that, along with RNA pol, are required for formation of the transcription initiation complex at the promoter. Although the TAD recruits a variety of proteins, the specific effect of any one of them is dependent upon the protein composition of the complex. This is known as combinatorial control.] Examples of coordinate regulation in eukaryotes include the galactose circuit and the hormone response system.
1. Galactose circuit: This regulatory scheme allows for the use of galactose when glucose is not available. In yeast, a unicellular organism, the genes required to metabolize galactose are on different chromosomes.
Coordinated expression is mediated by the protein Gal4 (Gal = galactose), a STF that binds to a short regulatory DNA sequence upstream of each of the genes. The sequence is called the upstream activating sequence Gal (UASGal). Binding of Gal4 to UASGal through zinc fingers in its DBD occurs in both the absence and presence of galactose. When the sugar is absent, the regulatory protein Gal80 binds Gal4 at its TAD, thereby inhibiting gene transcription (Fig.1A).
When present, galactose activates the Gal3 protein. Gal3 binds Gal80, thereby allowing Gal4 to activate transcription (Fig. 1B). [Note: Glucose prevents the use of galactose by inhibting expression of Gal4 protein.]

Figure 1: Regulation of galactose (gal) circuit in yeast in the A. absence and B. presence of galactose. [Note: Target genes, whether on the same or a different chromosome, each have an upstream activating sequence galactose (UASGal).] TAD = transcription activation domain; DBD = DNA-binding domain; mRNA = messenger RNA.
2. Hormone response system: Hormone response elements (HRE) are DNA sequences that bind trans-acting proteins and regulate gene expression in response to hormonal signals in multicellular organisms. Hormones bind to either intracellular (nuclear) receptors (for example, steroid hormones) or cell-surface receptors (for example, the peptide hormone glucagon).
a. Intracellular receptors: Members of the nuclear receptor superfamily, which includes the steroid hormone (glucocorticoids, mineralocorticoids, androgens, and estrogens), vitamin D, retinoic acid, and thyroid hormone receptors, function as STF. In addition to domains for DNA-binding and transcriptional activation, these receptors also contain a ligand-binding domain. For example, the steroid hormone cortisol (a glucocorticoid) binds intracellular receptors at the ligand-binding domain (Fig. 2). Binding causes a conformational change in the receptor that activates it.
The receptor–hormone complex enters the nucleus, dimerizes, and binds via a zinc finger motif to DNA at a regulatory element, the glucocorticoid response element (GRE) that is an example of a HRE. Binding allows recruitment of coactivators to the TAD and results in expression of cortisol-responsive genes, each of which is under the control of its own GRE. Binding of the receptor–hormone complex to the GRE allows coordinate expression of a group of target genes, even though these genes are on different chromosomes. The GRE can be located upstream or downstream of the genes it regulates and at great distances from them. The GRE, then, can function as a true enhancer . [Note: If associated with repressors, hormone–receptor complexes inhibit transcription.]
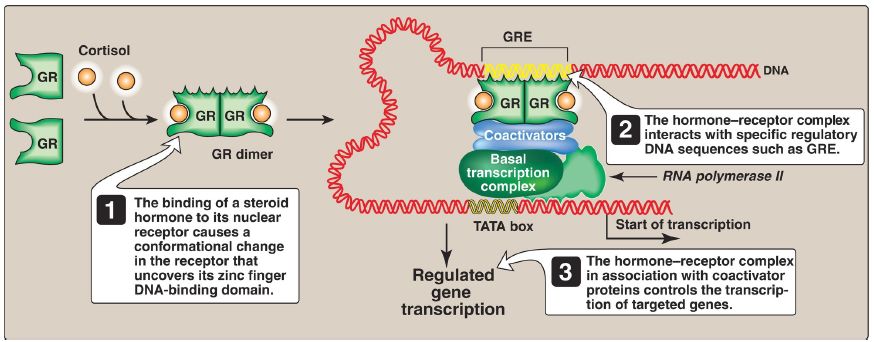
Figure 2: Transcriptional regulation by intracellular steroid hormone receptors. GRE = glucocorticoid response element; GR = glucocorticoid receptor.
b. Cell-surface receptors: These receptors include those for insulin, epinephrine, and glucagon. Glucagon, for example, is a peptide hormone that binds its G protein–coupled plasma membrane receptor on glucagon-responsive cells. This extracellular signal is then transduced to intracellular cAMP, a second messenger (Fig. 3 ), which can affect protein expression (and activity) through protein kinase A–mediated phosphorylation. In response to a rise in cAMP, a trans-acting factor (cAMP response element–binding [CREB] protein) is phosphorylated and activated. Active CREB protein binds via a leucine zipper motif to a cis-acting regulatory element, the cAMP response element (CRE), resulting in transcription of target genes with CRE in their promoters. [Note: The genes for phosphoenolpyruvate carboxykinase and glucose 6-phosphatase, key enzymes of gluconeogenesis are examples of genes upregulated by the cAMP/CRE/CREB system.]
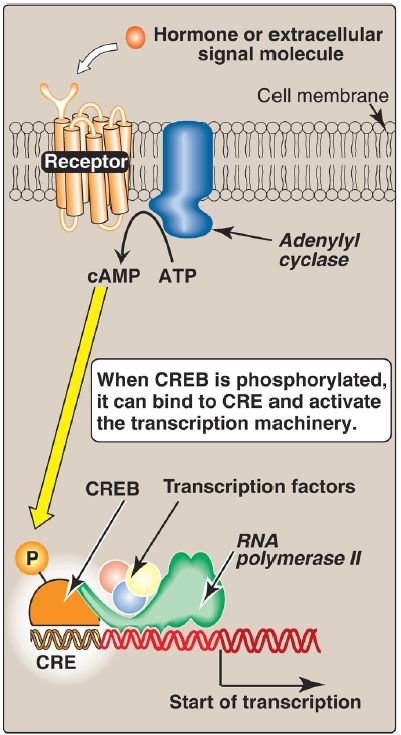
Figure 3: Transcriptional regulation by receptors located in the cell membrane. [Note: Cyclic adenosine monophosphate (cAMP) activates protein kinase A that phosphorylates cAMP response element–binding (CREB) protein.] CRE = cAMP response element.
 الاكثر قراءة في الكيمياء الحيوية
الاكثر قراءة في الكيمياء الحيوية
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)