النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Prokaryotic Gene Transcription : Steps in RNA synthesis
المؤلف:
Denise R. Ferrier
المصدر:
Lippincott Illustrated Reviews: Biochemistry
الجزء والصفحة:
26-12-2021
2149
Prokaryotic Gene Transcription : Steps in RNA synthesis
The process of transcription of a typical gene of Escherichia coli (E. coli) can be divided into three phases: initiation, elongation, and termination. A transcription unit extends from the promoter to the termination region, and the initial product of transcription by RNA pol is termed the primary transcript.
1. Initiation: Transcription begins with the binding of the RNA pol holoenzyme to a region of the DNA known as the promoter, which is not transcribed. The prokaryotic promoter contains characteristic consensus sequences (Fig. 1). [Note: Consensus sequences are idealized sequences in which the base shown at each position is the base most frequently (but not necessarily always) encountered at that position.] Those that are recognized by prokaryotic RNA pol σ factors include the following.
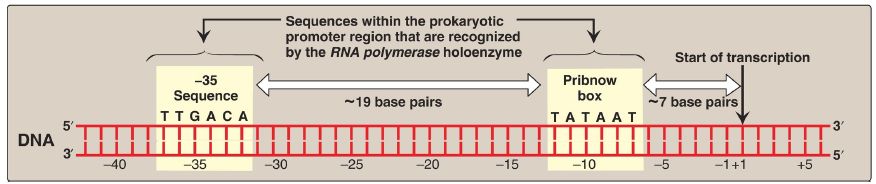
Figure 1: Structure of the prokaryotic promoter region. T = thymine; G = guanine; A = adenine; C = cytosine.
a. –35 Sequence: A consensus sequence (5′-TTGACA-3′), centered about 35 bases to the left of the transcription start site (see Fig. 1), is the initial point of contact for the holoenzyme, and a closed complex is formed. [Note: By convention, the regulatory sequences that control transcription are designated by the 5′→3′ nucleotide sequence on the coding strand. A base in the promoter region is assigned a negative number if it occurs prior to (to the left of, toward the 5′-end of, or “upstream” of) the transcription start site. Therefore, the TTGACA sequence is centered at approximately base −35. The first base at the transcription start site is assigned a position of +1. There is no base designated “0”.]
b. Pribnow box: The holoenzyme moves and covers a second consensus sequence (5′-TATAAT-3′), centered at about −10 (see Fig. 1), which is the site of melting (unwinding) of a short stretch (~14 base pairs) of DNA. This initial melting converts the closed initiation complex to an open complex known as a transcription bubble. [Note: A mutation in either the −10 or the −35 sequence can affect the transcription of the gene controlled by the mutant promoter.]
2. Elongation: Once the promoter has been recognized and bound by the holoenzyme, local unwinding of the DNA helix continues (Fig. 2), mediated by the polymerase. [Note: Unwinding generates supercoils in the DNA that can be relieved by DNA topoisomerases .]
RNA pol begins to synthesize a transcript of the DNA sequence, and several short pieces of RNA are made and discarded. The elongation phase begins when the transcript (typically starting with a purine) exceeds 10 nucleotides in length. Sigma is then released, and the core enzyme is able to leave (clear) the promoter and move along the template strand in a processive manner, serving as its own sliding clamp. During transcription, a short DNA–RNA hybrid helix is formed (see Fig. 2). Like DNA pol, RNA pol uses nucleoside triphosphates as substrates and releases pyrophosphate each time a nucleoside monophosphate is added to the growing chain. As with replication, transcription is always in the 5′→3′ direction. In contrast to DNA pol, RNA pol does not require a primer and does not have a 3′→5′ exonuclease domain for proofreading.
[Note: Misincorporation of a ribonucleotide causes RNA pol to pause, backtrack, cleave the transcript, and restart. Nonetheless, transcription has a higher error rate than does replication.]

Figure 2: Local unwinding of DNA by RNA polymerase and formation of an open initiation complex (transcription bubble).
3. Termination: The elongation of the single-stranded RNA chain continues until a termination signal is reached. Termination can be intrinsic (occur without additional proteins) or dependent upon the participation of a protein known as the ρ (rho) factor.
a. ρ-Independent: Seen with most prokaryotic genes, this requires that a sequence in the DNA template generates a sequence in the nascent (newly made) RNA that is self-complementary (Fig. 3). This allows the RNA to fold back on itself, forming a GC-rich stem (stabilized by hydrogen bonds) plus a loop. This structure is known as a “hairpin.” Additionally, just beyond the hairpin, the RNA transcript contains a string of Us at the 3′-end. The bonding of these Us to the complementary As of the DNA template is weak. This facilitates the separation of the newly synthesized RNA from its DNA template, as the double helix “zips up” behind the RNA pol.
Figure 3: Rho-independent termination of prokaryotic transcription. A. DNA template sequence generates a self-complementary sequence in the nascent RNA. B. Hairpin structure formed by the RNA. N represents a noncomplementary base; A = adenine, T = thymine; G = guanine; C = cytosine; U = uracil.
b. ρ-Dependent: This requires the participation of the additional protein rho, which is a hexameric ATPase with helicase activity. Rho binds a C-rich rho utilization (rut) site near the 5′-end of the nascent RNA and, using its ATPase activity, moves along the RNA until it reaches the RNA pol paused at the termination site. The ATP-dependent helicase activity of rho separates the RNA–DNA hybrid helix, causing the release of the RNA.
4. Antibiotics: Some antibiotics prevent bacterial cell growth by inhibiting RNA synthesis. For example, rifampin (rifampicin) inhibits transcription initiation by binding to the β subunit of prokaryotic RNA pol and preventing chain growth beyond three nucleotides (Fig. 4). Rifampin is important in the treatment of tuberculosis. Dactinomycin (actinomycin D) was the first antibiotic to find therapeutic application in tumor chemotherapy. It inserts (intercalates) between the DNA bases and inhibits transcription initiation and elongation.
Figure 4: Inhibition of prokaryotic RNA polymerase by rifampin (rifampicin).
 الاكثر قراءة في الكيمياء الحيوية
الاكثر قراءة في الكيمياء الحيوية
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)