


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 13-12-2015
Date: 20-11-2020
Date: 7-6-2021
|
A Riboswitch Can Alter Its Structure According to Its Environment
KEY CONCEPTS
- A riboswitch is an RNA whose activity is controlled by a small ligand (a ligand is any molecule that binds to another), which may be a metabolite product.
- A riboswitch may be a ribozyme.
As seen in the chapter titled The Operon, an mRNA is more than simply an open reading frame (ORF). Regions in the bacterial 5′ untranslated region (UTR) contain elements that, due to coupled transcription/translation, can control transcription termination. The 5′ UTR sequence itself can determine if an mRNA is a “good” message, which supports a high level of translation, or a “poor” message, which does not. Another type of element in a 5′ UTR that can control expression of the mRNA is a riboswitch. A riboswitch is an RNA domain that contains a sequence that can change in secondary structure to control its activity. This change can be mediated by small metabolites. It is important to note that RNA structural change can be at the level of secondary structure—how the RNA folds—or tertiary structure—how the RNA arms and loops associate together. These are independent structural features.
Dozens of different riboswitches have been identified, each responding to a different ligand. The RNA domain that binds the metabolite is called the aptamer. Aptamer binding causes a structural change to the platform, the remainder of the riboswitch that carries out its function. One type of riboswitch is an RNA element that can assume alternate base-pairing configurations (controlled by metabolites in the environment) that can affect translation of the mRNA. FIGURE .1 illustrates the regulation of the system that produces the metabolite GlcN6P (glucosamine-6-phosphate). The gene glmS codes for an enzyme that synthesizes GlcN6P from fructose-6-phosphate and glutamine. GlcN6P is a fundamental intermediate in cell wall biosynthesis in bacteria. The mRNA contains a long 5′ UTR before the coding region of the mRNA. (Extra-long 5′ or 3′ UTRs are a clue that there may be regulatory elements in them.) Within the 5′ UTR is a ribozyme—a sequence of RNA that has catalytic activity . In this case, the catalytic activity is an endonuclease that cleaves its own RNA. It is activated by binding of the metabolite product, GlcN6P, to the aptamer region of the ribozyme. The consequence is that accumulation of GlcN6P activates the ribozyme, which cleaves the mRNA, which, in turn, prevents further translation. This is an exact parallel to allosteric control of a repressor protein by the end product of a metabolic pathway. There are numerous examples of such riboswitches in bacteria.
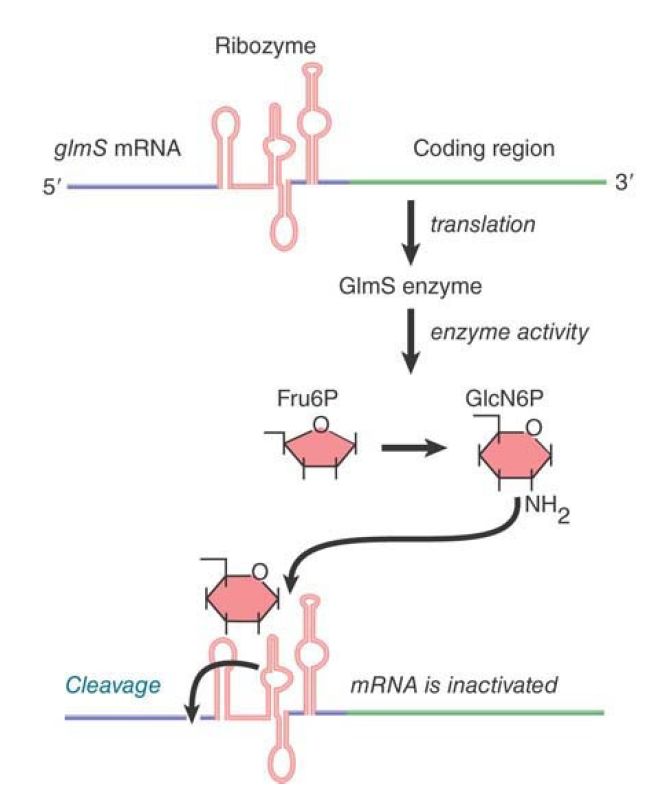
FIGURE .1 The 5′ untranslated region of the mRNA for the enzyme that synthesizes GlcN6P contains a ribozyme that is activated by the metabolic product. The ribozyme inactivates the
mRNA by cleaving it.
Not all riboswitches encode a ribozyme that controls mRNA stability. Other riboswitches have alternate configurations of the RNA that allow or prevent expression of the mRNA by affecting
ribosome binding. Riboswitches are found predominantly in bacteria and less commonly in eukaryotes.
An interesting eukaryotic riboswitch has been described in the fungus Neurospora to control alternate splicing. The gene NMT1 (involved with vitamin B1 synthesis) produces an mRNA precursor with a single intron that has two splice donor sites . Alternative use of these two sites can produce a functional or nonfunctional message depending on the concentration of a vitamin B1 metabolite, thiamine pyrophosphate (TPP). Thus, product concentration controls product formation, a form of repressible control. The selection of the splice site is controlled by a riboswitch in the intron. At a low concentration of TPP the proximal splice donor site is chosen and the distal splice donor site is blocked by the riboswitch, as shown in FIGURE .2. This splice produces a functional mRNA. At high TPP concentration, TPP binds the riboswitch to alter its configuration and prevents blocking of the distal splice donor site to allow the alternate splice, which produces a nonfunctional mRNA.
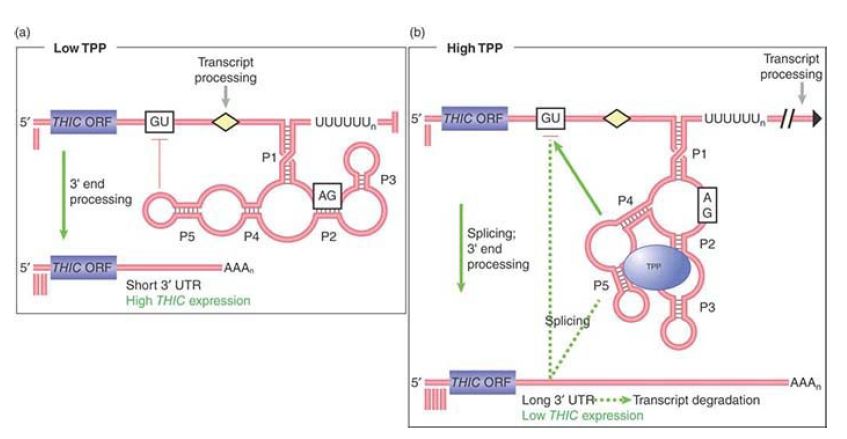
FIGURE .2 Expression of the NMT1 gene is regulated at the level of pre-mRNA alternate splicing by a riboswitch that binds to TPP. (a) At low concentrations of TPP, the TPP-binding aptamer region of the riboswitch base pairs with sequences surrounding a splice site (red blocking line) in a nearby noncoding sequence and prevents its selection by the splicing machinery. A distal splice site is selected resulting in a short mRNA with an open reading frame (ORF) that translates into a functional protein. (b) At high TPP levels, the aptamer undergoes a conformational rearrangement so that the region that was previously bound to the nearby splice site is now bound to TPP. This and other conformational changes results in a longer mRNA splice variant that contains short decoy ORFs, preventing functional NMT1 expression.
Reproduced from A. Wachter, et al. Plant Cell 19 (2007): 3437–3450.



|
|
|
|
مخاطر خفية لمكون شائع في مشروبات الطاقة والمكملات الغذائية
|
|
|
|
|
|
|
"آبل" تشغّل نظامها الجديد للذكاء الاصطناعي على أجهزتها
|
|
|
|
|
|
|
تستخدم لأول مرة... مستشفى الإمام زين العابدين (ع) التابع للعتبة الحسينية يعتمد تقنيات حديثة في تثبيت الكسور المعقدة
|
|
|