
tRNAs Are Processed from Longer Precursors
 المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
 المصدر:
LEWIN’S GENES XII
المصدر:
LEWIN’S GENES XII
 الجزء والصفحة:
الجزء والصفحة:
 29-5-2021
29-5-2021
 2279
2279
tRNAs Are Processed from Longer Precursors
KEY CONCEPTS
- A mature tRNA is generated by processing a precursor.
- The 5′ end is generated by cleavage by the endonuclease RNase P.
- The 3′ end is generated by multiple endonucleolytic and exonucleolytic cleavages, followed by addition of the common terminal trinucleotide CCA.
tRNAs are commonly synthesized as precursor chains with additional sequences at one or both ends. FIGURE 1 shows that the extra sequences are removed by combinations of endonucleolytic and exonucleolytic activities. The three nucleotides at the 3′ terminus, which are always present as the triplet sequence CCA, are sometimes not encoded in the genome. In such cases, they are added as part of the tRNA processing.
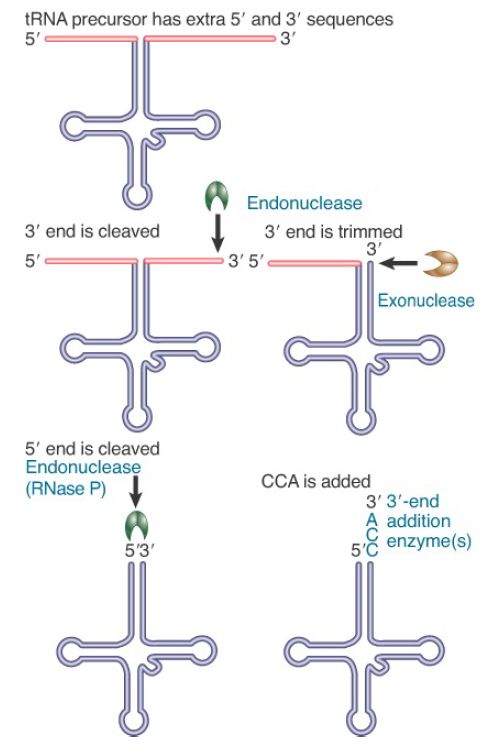
FIGURE 1. The tRNA 3′ end is generated by cutting (endonucleolytic) and trimming (exonucleolytic) reactions, followed by addition of CCA when this sequence is not encoded; the 5′ end is generated by a precise endonucleolytic cleavage.
The 5′ end of tRNA is generated by a cleavage action catalyzed by the ribonucleoprotein enzyme ribonuclease P. This enzyme recognizes the global L-shaped tRNA structure and specifically hydrolyzes the phosphodiester linkage that forms the mature 5′ end of the molecule, leaving a 5′-phosphate group. In E. coli, RNase P consists of a 377-nucleotide RNA and 17.5-kD protein, and its active site is composed of RNA. In vitro the RNA component alone is able to catalyze the tRNA-processing reaction. The function of the protein subunit is to stabilize a conformation of the RNA active site that is complementary to the tRNA precursor.
In the case of histidine-specific tRNAs in some organisms, after RNase P cleavage an additional guanosine residue is added at the 5′ terminus, thus forming a unique G-1 nucleotide. The enzyme that accomplishes this addition, Thg1, has the remarkable property of catalyzing the equivalent of a reverse polymerization reaction. The new guanosine is added by nucleotide addition in the 3′ to 5′ direction, opposite to that of all other known DNA and RNA polymerases.
The enzymes that process the 3′ end are best characterized in E. coli, where an endonuclease triggers the reaction by cleaving the precursor downstream, and several exonucleases then trim the end by degradation in the 3′ to 5′ direction. tRNA 3′-end processing also involves several enzymes in eukaryotes. The addition of the 3′-CCA is catalyzed by the enzyme tRNA nucleotidyltransferase, which functions as a non-template-directed RNA polymerase; that is, the enzyme specifically adds C, C, and A in sequence, without pairing the cytosine and adenine to complementary guanine and uracil bases on a template. Instead, the enzyme structure itself is sufficient to form sequential complementary binding sites for C, C, and A. As the nucleotides are added, the enzyme–tRNA complex changes conformation to become complementary to each successive nucleotide.
All three nucleotides are added by tRNA nucleotidyltransferase when they are not encoded in the tRNA gene sequence. Interestingly, the enzyme also plays an essential role in repairing damaged tRNA 3′ ends in organisms such as E. coli that do encode CCA. In these organisms, three different tRNA substrates are recognized: those lacking CCA, those possessing a 3′-C, and those possessing a 3′-CC.
tRNA nucleotidyltransferase enzymes are divided into two classes that retain significant amino acid similarity only in their active site regions. Class I enzymes are found in archaea; bacterial and eukaryotic enzymes together make up a second class. In some very ancient bacterial lineages, CCA addition is catalyzed by two closely related class II enzymes: one of these enzymes adds –CC, and the other adds the 3′-terminal A.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


