


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 26-4-2016
Date: 25-12-2015
Date: 15-11-2020
|
Viroids Have Catalytic Activity
KEY CONCEPTS
- Viroids and virusoids form a hammerhead structure that has a self-cleaving activity.
- Similar structures can be generated by pairing a substrate strand that is cleaved by an enzyme strand.
- When an enzyme strand is introduced into a cell, it can pair with a substrate strand target that is then cleaved.
Another example of the ability of RNA to function as an endonuclease is provided by some small plant RNAs of about 350 nucleotides that undertake a self-cleavage reaction. However, as with the case of the Tetrahymena group I intron, it is possible to engineer constructs that can function on external substrates.
These small plant RNAs fall into two general groups: viroids and virusoids. The viroids are infectious RNA molecules that function independently without encapsidation by any protein coat. The virusoids (which are sometimes called satellite RNAs) are similar in organization but are encapsidated by plant viruses, being packaged together with a viral genome. The virusoids cannot replicate independently; they require assistance from the virus.
Viroids and virusoids both replicate via rolling circles. The strand of RNA that is packaged into the virus is called the plus strand. The complementary strand, generated during replication of the RNA, is called the minus strand. Multimers of both plus and minus strands are found. Both types of monomer are generated by cleaving the tail of a rolling circle; circular plus-strand monomers are generated by ligating the ends of the linear monomer.
Both plus and minus strands of viroids and virusoids undergo selfcleavage in vitro. Some of the RNAs cleave in vitro under physiological conditions. Others do so only after a cycle of heating and cooling; this suggests that the isolated RNA has an inappropriate conformation, but can generate an active conformation when it is denatured and renatured. The viroids and virusoids that undergo self-cleavage form a “hammerhead” secondary structure at the cleavage site, as shown in the upper part of FIGURE 1. Hammerhead ribozymes belong to a family of ribozymes that includes hepatitis delta virus (HDV), hairpin ribozymes, and Varkud satellite (VS) ribozyme.
Functionally, HDV requires divalent metal cations to promote cleavage, whereas hammerhead and hairpin ribozymes do not require metal. The importance of metal for VS ribozyme cleavageis still ambiguous . However, all of these ribozymes generate a cleavage that leaves 5′–OH and 2′,3′-cyclic phosphodiester termini.
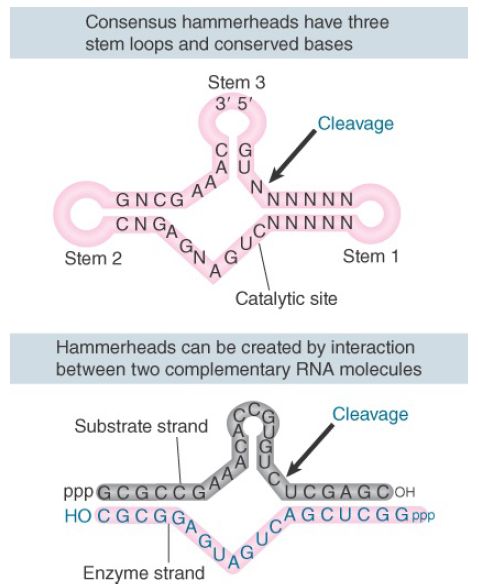
FIGURE 1. Self-cleavage sites of viroids and virusoids have a consensus sequence and form a hammerhead secondary structure by intramolecular pairing. Hammerheads can also be generated by pairing between a substrate strand and an “enzyme” strand. The three loop regions at the end of the stems are optional.
The number of hammerhead ribozymes identified now exceeds 10,000, with examples found in all three taxonomic domains. Unlike all other ribozymes identified to date, hammerhead ribozymes and other members of the family do not require a protein component to function in vivo because the sequence of this structure is sufficient for cleavage. Minimally, for hammerhead ribozymes the active site is a sequence of only 58 nucleotides. The hammerhead contains three stem-loop regions whose positions and sizes are constant and 13 conserved nucleotides, mostly in the regions connecting the center of the structure. Hammerhead ribozymes can be further divided into classes I, II, and III, corresponding to the stem in which the free 5′ and 3′ ends of the RNA reside. The conserved bases and duplex stems generate an RNA with the intrinsic ability to cleave.
An active hammerhead can also be generated by pairing an RNA representing one side of the structure with an RNA representing the other side. The lower part of Figure 1 shows an example of ahammerhead generated by h ybridizing a 19-nucleotide molecule with a 24-nucleotide molecule. The hybrid mimics the hammerhead structure, with the omission of loops I and III. We may regard the top (24-nucleotide) strand of this hybrid as comprising the “substrate” and the bottom (19-nucleotide) strand as comprising the “enzyme.” When the 19-nucleotide RNA is added to the 24-nucleotide RNA, cleavage occurs at the appropriate position in the hammerhead. When the 19-nucleotide RNA is mixed with an excess of the 24-nucleotide RNA, multiple copies of the 24-nucleotide RNA are cleaved. This suggests that there is a cycle of 19-nucleotide to 24-nucleotide pairing, cleavage, dissociation of the cleaved fragments from the 19-nucleotide RNA, and pairing of the 19-nucleotide RNA with a new 24-nucleotide substrate. The 19-nucleotide RNA is therefore a ribozyme with endonuclease activity. The parameters of the reaction are similar to those of other RNAcatalyzed reactions.
Previously, the crystal structure of a minimal hammerhead ribozyme was solved. However, in the minimal structure, the architecture of the active site was such that it was unclear how catalysis proceed. More recently, the crystal structure of the full-length hammerhead ribozyme from Schistosoma mansoni, a nonvirulent species, has been solved, and it gives insight into catalysis. This structure, schematically illustrated in FIGURE 2, reveals a critical tertiary interaction between a bulge in stem I and the loop of stem II. This interaction stabilizes the active site in a conformation such that G12 can deprotonate the 2′–OH of C17 and the scissile bond and create the 2′-attacking oxygen. In turn, G8 provides the hydrogen to stabilize the newly formed 5′–OH end of the 3′ cleavage product.
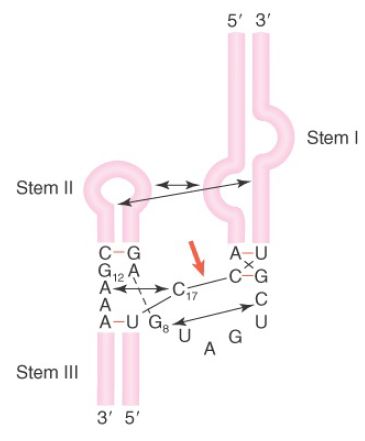
FIGURE 2. The hammerhead ribozyme structure is held in an active tertiary conformation by interactions between stem-loops, indicated by arrows. The site of cleavage is marked with a red arrow.
Data from M. Martick and W. G. Scott, Cell (126): 309–320.
It is possible to design enzyme–substrate combinations that can form minimal hammerhead structures. These structures have been used to demonstrate that introduction of the appropriate RNA molecules into a cell can allow the enzymatic reaction to occur in vivo. A ribozyme designed in this way essentially provides a highly specific restriction endonuclease-like activity directed against an RNA target. By placing the ribozyme under control of a regulated promoter, it can be used in the same way as, for example, antisense constructs to specifically turn off expression of a target gene under defined circumstances.



|
|
|
|
التوتر والسرطان.. علماء يحذرون من "صلة خطيرة"
|
|
|
|
|
|
|
مرآة السيارة: مدى دقة عكسها للصورة الصحيحة
|
|
|
|
|
|
|
نحو شراكة وطنية متكاملة.. الأمين العام للعتبة الحسينية يبحث مع وكيل وزارة الخارجية آفاق التعاون المؤسسي
|
|
|