


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 18-5-2016
Date: 8-4-2021
Date: 30-11-2015
|
Some Eukaryotic mRNAs Are Localized to Specific Regions of a Cell
KEY CONCEPTS
- Localization of mRNAs serves diverse functions in single cells and developing embryos.
- Three mechanisms for the localization of mRNA have been documented.
- Localization requires cis-elements on the target mRNA and trans-factors to mediate the localization.
- The predominant active transport mechanism involves the directed movement of mRNPs along cytoskeletal tracks.
The cytoplasm is a crowded place occupied by a high concentration of proteins. It is not clear how freely polysomes can diffuse, and most mRNAs are probably translated in random locations that are determined by their point of entry into the cytoplasm and the distance that they may have moved away from it. Some mRNAs are translated only at specific sites, though—their translation is repressed until they reach their destinations. The regulated localization has been described for more than 100 specific mRNAs, a number that certainly represents a small fraction of the total. mRNA localization serves a number of important functions in eukaryotic organisms of all types. Three key functions are illustrated in FIGURE 1 and described below:
1. Localization of specific mRNAs in the oocytes of many animals serves to set up future patterns in the embryo (such as axis polarity) and to assign developmental fates to cells residing in different regions. These localized maternal mRNAs encode transcription factors or other proteins that regulate gene expression. In Drosophila oocytes, bicoid and nanos mRNAs are localized to the anterior and posterior poles, respectively, and their translation following fertilization results in gradients of their protein products. The gradients are used by cells in early development for the specification of their anterior–posterior position in the embryo. Bicoid encodes a transcription factor, and nanos encodes a translational repressor. Some localized mRNAs encode determinants of cell fate. For example, oskar mRNA localizes in the posterior of the oocyte and initiates the process leading to development of primordial germ cells in the embryo. It is estimated that during Drosophila development 70% of mRNAs are expressed in specific spatial domains.
2. mRNA localization also plays a role in asymmetric cell divisions; that is, mitotic divisions that result in daughter cells that differ from one another. One way this is accomplished is by asymmetric segregation of cell-fate determinants, which may be proteins and/or the mRNAs that encode them. In Drosophila embryos, prospero mRNA and its product (a transcription factor) are localized to a region of the peripheral cortex of the embryo. Later in development, oriented cell division of neuroblasts ensures that only the outermost daughter cell receives prospero, committing it to a ganglion mother-cell fate. Asymmetric cell division is also used by budding yeast to generate a daughter cell of a different mating type than the mother cell, an event described later in this section.
3. mRNA localization in adult, differentiated cell types is a mechanism for the compartmentalization of the cell into specialized regions. Localization may be used to ensure that components of multiprotein complexes are synthesized in proximity to one another and that proteins targeted to organelles or specialized areas of cells are synthesized conveniently nearby. mRNA localization is particularly important for highly polarized cells such as neurons. Although most mRNAs are translated in the neuron cell body, many mRNAs are localized to its dendritic and axonal extensions. Among those is β-actin mRNA, whose product participates in dendrite and axon growth. β-actin mRNA localizes to sites of active movement in a wide variety of motile cell types. Interestingly, localization of mRNA at neuronal postsynaptic sites seems to be essential for modifications accompanying learning. In glial cells, the myelin basic protein (MBP) mRNA, which encodes a component of the myelin sheath, is localized to a specific myelin-synthesizing compartment. Plants localize mRNAs to the cortical region of cells and to regions of polar cell growth.

FIGURE 1.Three main functions of mRNA localization.
In some cases, mRNA localization involves transport from one cell to another. Maternal mRNPs in Drosophila are synthesized and assembled in surrounding nurse cells and are transferred to the developing oocyte through cytoplasmic canals. Plants can export RNAs through plasmodesmata and transport them for long distances via the phloem vascular system. mRNAs are sometimes transported en masse in mRNP granules. The compositions of these granules are not yet well defined.
Three mechanisms for the localization of mRNA have been well documented:
1. The mRNA is uniformly distributed but degraded at all sites except the site of translation.
2. The mRNA is freely diffusible but becomes trapped at the site of translation.
3. The mRNA is actively transported to a site where it is translated.
Active transport is the predominant mechanism for localization. Transport is achieved by translocation of motor proteins along cytoskeletal tracks. All three molecular motor types are exploited: dyneins and kinesins, which travel along microtubules in opposite directions, and myosins, which travel along actin fibers. This mode of localization requires at least four components: (1) cis-elements on the target mRNA, (2) trans-factors that directly or indirectly attach the mRNA to the correct motor protein, (3) trans-factors that repress translation, and (4) an anchoring system at the desired location.
Only a few cis-elements, sometimes called zipcodes, have been characterized. They are diverse, include examples of both sequence and structural RNA elements, and can occur anywhere in the mRNA, though most are in the 3′ UTR. Zipcodes have been difficult to identify, presumably because many consist of complex secondary and tertiary structures. A large number of trans-factors have been associated with localized mRNA transport and translational repression, some of which are highly conserved in different organisms. For example, staufen, a double-stranded RBP, is involved in localizing mRNAs in the oocytes of Drosophila and Xenopus, as well as the nervous systems of Drosophila, mammals, and probably worms and zebrafish. This multitalented factor has multiple domains that can couple complexes to both actin- and microtubule-dependent transport pathways. Almost nothing is known about the fourth required component—anchoring mechanisms. Two examples of localization mechanisms are discussed in the following paragraphs.
The localization of β-actin mRNA has been studied in cultured fibroblasts and neurons. The zipcode is a 54-nucleotide element in the 3′ UTR. Cotranscriptional binding of the zipcode element by the protein ZBP1 is required for localization, suggesting that this mRNA is committed to localization before it is even processed and exported from the nucleus. Interestingly, β-actin mRNA localization is dependent on intact actin fibers in fibroblasts and intact microtubules in neurons.
Genetic analysis of ASH1 mRNA localization in yeast has provided the most complete picture of a localization mechanism to date and is illustrated in FIGURE 2 . During budding, the ASH1 mRNA is localized to the developing bud tip, resulting in Ash1 synthesis only in the newly formed daughter cell. Ash1 is a transcriptional repressor that disallows expression of the HO endonuclease, a protein required for mating-type switching . The result is that mating-type switching occurs only in the mother cell. The ASH1 mRNA has four stem-loop localization elements in its coding region to which the protein She2 binds, probably in the nucleus. The protein She3 serves as an adaptor, binding both to She2 and to the myosin motor protein Myo4 (also called She1). A Puf protein, Puf6, binds to the mRNA, repressing its translation. The motor transports the ASH1 mRNP along the polarized actin fibers that lead from the mother cell to the developing bud. Additional proteins are required for proper localization and expression of the ASH1 mRNA. More than 20 yeast mRNAs use the same localization pathway.
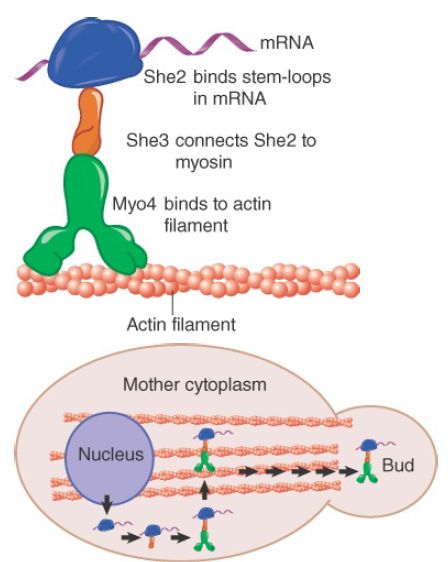
FIGURE 2. Localization of ASH1 mRNA. Newly exported ASH1 mRNA is attached to the myosin motor Myo4 via a complex with the She2 and She3 proteins. The motor transports the mRNA along actin filaments to the developing bud.
Localization mechanisms that do not involve active transport have been clearly demonstrated for only a few localized mRNAs in oocytes and early embryos. The mechanism of local entrapment of diffusible mRNAs requires the participation of previously localized anchors, which have not been identified. In Drosophila oocytes, diffusing nanos mRNA is trapped at the posterior germ plasm, a specialized region of the cytoplasm underlying the cortex. In Xenopus oocytes, mRNAs localized to the vegetal pole are first trapped in a somewhat mysterious, membrane-laden structure called the mitochondrial cloud (MC), which later migrates to the vegetal pole, carrying mRNAs with it. The mechanism of localized mRNA stabilization has been described for an mRNA that also localizes to the posterior pole of the Drosophila embryo. Early in development, the hsp83 mRNA is uniformly distributed through the embryonic cytoplasm, but later it is degraded everywhere except at the pole. A protein called smaug is involved in destabilizing the majority of the hsp83 mRNAs, most likely by recruiting the CCR4-NOT complex. How the pole-localized mRNAs escape is not known.



|
|
|
|
مخاطر خفية لمكون شائع في مشروبات الطاقة والمكملات الغذائية
|
|
|
|
|
|
|
"آبل" تشغّل نظامها الجديد للذكاء الاصطناعي على أجهزتها
|
|
|
|
|
|
|
تستخدم لأول مرة... مستشفى الإمام زين العابدين (ع) التابع للعتبة الحسينية يعتمد تقنيات حديثة في تثبيت الكسور المعقدة
|
|
|