


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 31-10-2020
Date: 10-12-2020
Date: 30-4-2021
|
TBP Is a Universal Factor
KEY CONCEPTS
- TATA-binding protein (TBP) is a component of the positioning factor that is required for each type of RNA polymerase to bind its promoter.
- The factor for RNA polymerase II is TFIID, which consists of TBP and about 14 TBP-associated factors (TAFs), with a total mass of about 800 kD.
- TBP binds to the TATA box in the minor groove of DNA.
- TBP forms a saddle around the DNA and bends it by approximately 80°.
Before transcription initiation can begin, the chromatin has to be modified and remodeled to the open configuration, and any nucleosome octamer positioned over the promoter has to be moved or removed at all classes of eukaryotic promoters (we examine this aspect of transcription control more closely in the chapter titled Eukaryotic Transcription Regulation). At that point it is possible for a positioning factor to bind to the promoter. Each class of RNA polymerase is assisted by a positioning factor that contains TBP associated with other components. Recall that TBP stands for TATA-binding protein; it was initially so named because it was a protein that bound to the TATA box in RNA polymerase II genes. It was subsequently discovered to also be part of the positioning factors SL1 for RNA polymerase I and TF B RNA polymerase III .
For these latter two RNA polymerases, TBP does not recognize the TATA box sequence (except in type 3 pol III promoters); thus, the name is misleading. In addition, many RNA polymerase II promoters lack TATA boxes, but still require the presence of TBP.
For RNA polymerase II, the positioning factor is TFII D, which consists of TBP associated with up to 14 other subunits called TAFs (for TBP-associated factors). Some TAFs are stoichiometric with TBP; others are present in lesser amounts, which means that there are multiple TFII D variants. TFII Ds containing different TAFs could recognize promoters with different combinations of conserved elements described in the previous section, The Start Point for RNA Polymerase II. Some TAFs are tissue specific. The total mass of TFII D typically is about 800 kD. The TAFs in TFII D were originally named in the form TAF 00, for example, where the number 00 gives the molecular mass of the subunit. Recently, the RNA polymerase II TAFs have been renamed TAF1, TAF2, and so forth; in this nomenclature TAF1 is the largest TAF, TAF2 is the next largest, and homologous TAFs in different species thus have the same names.
FIGURE 1 shows that the positioning factor recognizes the promoter in a different way in each case. At promoters for RNA polymerase III, TFIII B binds adjacent to TFIII C. At promoters for RNA polymerase I, SL1 binds in conjunction with UBF. TFII D is solely responsible for recognizing promoters for RNA polymerase II. At a promoter that has a TATA element, TBP binds specifically to the TATA box, but at TATA-less promoters, the TAFs have the role of recognizing other promoter elements, including the Inr and DPE. Whatever its means of entry into the initiation complex, it has the common purpose of interaction with the RNA polymerase.

FIGURE 1. RNA polymerases are positioned at all promoters by a factor that contains TBP.
TBP has the unusual property of binding to DNA in the minor groove. (The vast majority of DNA-binding proteins bind in the major groove.) The crystal structure of TBP suggests a detailed model for its binding to DNA. FIGURE 2 shows that it surrounds one face of DNA, forming a “saddle” around a stretch of the minor groove, which is bent to fit into this saddle. In effect, the inner surface of TBP binds to DNA, and the larger outer surface is available to extend contacts to other proteins. The DNA-binding site consists of a C-terminal domain that is conserved between species, and the variable N-terminal tail is exposed to interact with other proteins. It is a measure of the conservation of mechanism in transcriptional initiation that the DNA-binding sequence of TBP is 80% conserved between yeast and humans.
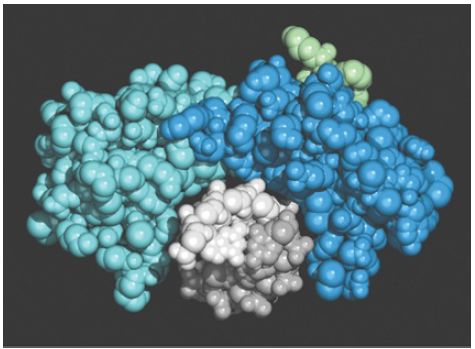
FIGURE 2. A view in cross-section shows that TBP surrounds DNA from the side of the narrow groove. TBP consists of two related (40% identical) conserved domains, which are shown in light and dark blue. The N-terminal region varies extensively and is shown in green. The two strands of the DNA double helix are in light and dark gray.
Photo courtesy of Stephen K. Burley.
Binding of TBP may be inconsistent with the presence of nucleosome octamers. Nucleosomes form preferentially by placing A-T−rich sequences with the minor grooves facing inward ; as a result, they could prevent binding of TBP. This may explain why the presence of a nucleosome at the promoter prevents initiation of transcription.
TBP binds to the minor groove and bends the DNA by approximately 80°, as illustrated in FIGURE 2. The TATA box bends toward the major groove, widening the minor groove. The distortion is restricted to the 8 bp of the TATA box; at each end of the sequence the minor groove has its usual width of about 5 Å, but at the center of the sequence the minor groove is greater than 9 Å. This is a deformation of the structure, but it does not actually separate the strands of DNA because base pairing is maintained. The extent of the bend can vary with the exact sequence of the TATA box and is correlated with the efficiency of the promoter.
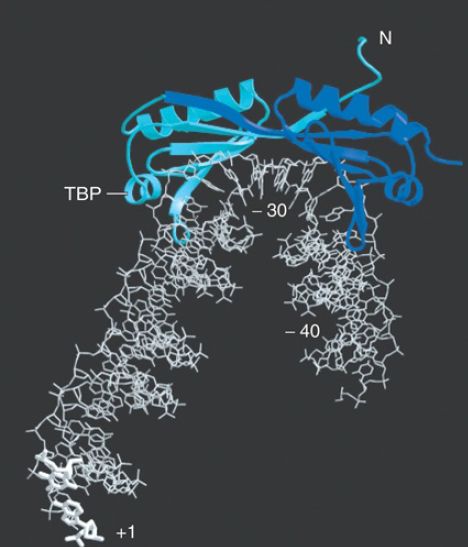
FIGURE 18.10 The cocrystal structure of TBP with DNA from −40 to the start point shows a bend at the TATA box that widens the narrow groove where TBP binds.
Photo courtesy of Stephen K. Burley.
This structure has several functional implications. By changing the spatial organization of DNA on either side of the TATA box, it allows the transcription factors and RNA polymerase to form a closer association than would be possible on linear DNA. The bending at the TATA box corresponds energetically to unwinding of about onethird of a turn of DNA, and is compensated by a positive writhe.
The presence of TBP in the minor groove, combined with other proteins binding in the major groove, creates a high density of protein–DNA contacts in this region. Binding of purified TBP to DNA in vitro protects about one turn of the double helix at the TATA box, typically extending from −37 to −25. Binding of the TFII D complex in the initiation reaction, however, regularly protects the region from −45 to −10.
Within TF D as a free protein complex, the factor TAF1 binds to TBP, where it occupies the concave DNA-binding surface. In fact, the structure of the binding site, which lies in the N-terminal domain of TAF1, mimics the surface of the minor groove in DNA. This molecular mimicry allows TAF1 to control the ability of TBP to bind to DNA; the N-terminal domain of TAF1 must be displaced from the DNA-binding surface of TBP in order for TFII D to bind to DNA. Strikingly, a number of TAFs resemble histones: 9 of 14 TAFs contain a histone fold domain, though in most cases the TAFs lack the residues of this domain that are responsible for DNA binding.
Four TAFs do have some intrinsic DNA binding ability: TAF4b, TAF12, TAF9, and TAF6 are (distant) homologs of histones H2A, H2B, H3, and H4, respectively. (The histones form the basic complex that binds DNA in eukaryotic chromatin; see the chapter titled Chromatin.)
TAF4b/TAF12 and TAF9/TAF6 form heterodimers using the histone-fold motif; together they may form the basis for a structure resembling a histone octamer. Such a structure may be responsible for non-sequence-specific interactions of TFII D with DNA. Histone folds are also used in pairwise interactions between other TAF s.
Some of the TAF s may be found in other complexes as well as in TFII D. In particular, the histone-like TAF s also are found in protein complexes that modify the structure of chromatin prior to transcription (see the chapter titled Eukaryotic Transcription Regulation).



|
|
|
|
تفوقت في الاختبار على الجميع.. فاكهة "خارقة" في عالم التغذية
|
|
|
|
|
|
|
أمين عام أوبك: النفط الخام والغاز الطبيعي "هبة من الله"
|
|
|
|
|
|
|
قسم شؤون المعارف ينظم دورة عن آليات عمل الفهارس الفنية للموسوعات والكتب لملاكاته
|
|
|