النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Antitermination Can Be a Regulatory Event
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
6-5-2021
3086
Antitermination Can Be a Regulatory Event
KEY CONCEPTS
- An antitermination complex allows RNA polymerase to read through terminators.
- Phage lambda uses antitermination systems for regulation of both its early and late transcripts, but the two systems work by completely different mechanisms.
- Binding of factors to the nascent RNA links the antitermination proteins to the terminator site through an RNA loop.
- Antitermination of transcription also occurs in rRNA operons.
Antitermination is used as a mechanism for control of transcription in both phage and bacterial operons. As shown in FIGURE 1, antitermination refers to modification of the enzyme, which allows it to read past a terminator into genes that lie downstream. In the example shown in the figure, the default pathway is for RNA polymerase to terminate at the end of region 1, but antitermination results in continued transcription through region 2.

FIGURE 1.Antitermination can control transcription by determining whether RNA polymerase terminates or reads through a particular terminator into the following region.
Antitermination systems are common in lambdoid bacteriophages (phages similar to phage lambda, described in the chapter titled Phage Strategies). Unlike the E. coli T7-like phages and the B. subtilis SPO1 phages discussed earlier, lambda does not encode either its own dedicated RNA polymerase or even its own dedicated sigma factors. Rather, it uses the host multisubunit RNA polymerase for all of its transcription. Shortly after phage infection, transcription begins at two early promoters, PR and PL. However, terminators in each of these operons follow the transcription start site before most of the genes that encode most early functions, and termination of transcription at these positions aborts the infection. If RNA polymerase reads through the terminators and transcribes the early genes responsible for replication of the phage
genome, though, lambda development proceeds.
The first termination decision is controlled by an antitermination protein called N, which is the first protein produced by expression from PL . N forms a complex with host proteins called Nus factors (N utilization substances) to modify RNA polymerase in such a way that it no longer responds to the terminators. The antitermination complex actually forms on the nascent RNA at a sequence called nut (N utilization site). nut sites consist primarily of RNA sequences called boxA and boxB where the host factors NusA, NusB, NusE (ribosomal protein S10), and NusG assemble. The antitermination proteins remain bound to these RNA sites as a persistent antitermination complex as RNA polymerase synthesizes the two transcripts to the right and the left. Thus, the nascent RNA physically connects the antitermination proteins bound to the nut site with the RNA polymerase as it approaches terminators.
Although the actual mechanism by which the antitermination complex prevents termination is still not understood, tethering of the antitermination proteins to RNA polymerase through the nascent RNA explains its ability to antiterminate at successive terminators spaced hundreds or even thousands of bases downstream. The last protein produced by the N-antiterminated transcript from the other early promoter, PR , is named Q. Like N, Q is an antitermination protein. Q antiterminates transcription from the late promoter P , which produces a transcript coding for the phage’s head and tail proteins. Thus, lambda gene expression occurs in two stages, each of which is controlled by antitermination (see the chapter titled Phage Strategies and FIGURE 2). Q enables RNA polymerase to read through terminators in the late transcription unit, but it does so by a completely different mechanism than N. Unlike N, Q binds DNA (at the qut, Q utilization, site), but like N it travels with RNA polymerase and somehow interferes with the action of terminators throughout the late operon.
It appears that the action of Q involves acceleration of RNA polymerase through pause sites. (We discuss the overall regulation of lambda development in the chapter titled Phage Strategies.)
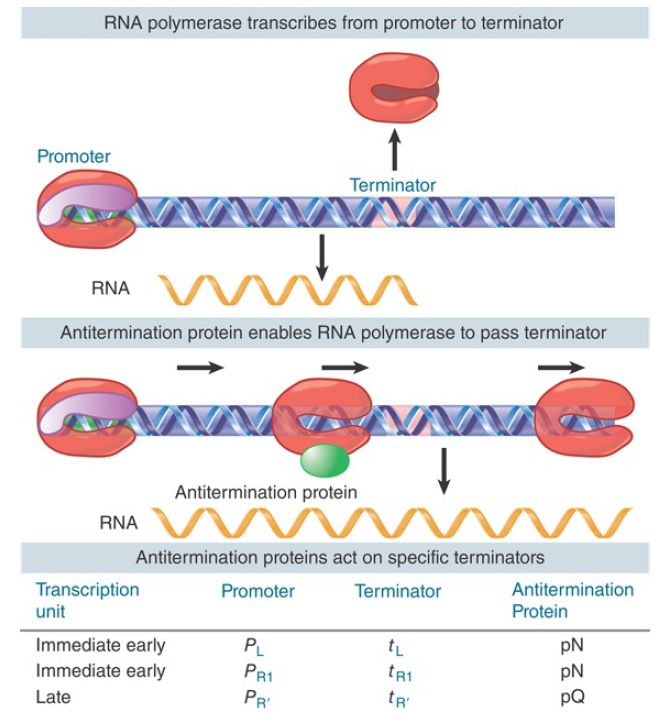
FIGURE 2. An antitermination protein can act on RNA polymerase to enable it to read through a specific terminator.
rRNA operons might be expected to exhibit polarity, because they are long but are not translated. Each of the rRNA operons of E. coli, however, contains boxA- and boxB-like sequences that assemble antitermination complexes on the transcripts consisting of at least some of the same Nus factors as those utilized by phage lambda. These complexes do not contain an N- or Q-like factor, which are encoded only by phage genomes, but they are sufficient to prevent premature termination at the hairpin sequences and weak rho-dependent terminators that occur fortuitously within the rRNA structural genes. Antitermination is needed for efficient rRNA production all the time, not just when lambda infects cells. Thus, bacterial evolution did not select for the Nus factors to facilitate lambda gene expression. Rather, these factors undoubtedly evolved to prevent polarity in rRNA operons. The leader regions of the rrn operons contain boxA sequences that assemble the Nus factors as the boxA sequences in RNA emerge from the RNA exit channel. As with antitermination in lambda, this process somehow changes the properties of RNA polymerase in such a way that it can now read through terminators, although the mechanism remains unclear.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)