النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Methylation of the Bacterial Origin Regulates Initiation
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
1-4-2021
2319
Methylation of the Bacterial Origin Regulates Initiation
KEY CONCEPTS
- oriC contains binding sites for DnaA: dnaA boxes.
- oriC also contains 11 repeats that are methylated on adenine on both strands.
- Replication generates hemimethylated DNA, which cannot initiate replication.
- There is a 13-minute delay before the repeats are remethylated.
The bacterial DnaA protein is the replication initiator; it binds sequence specifically to multiple sites (dnaA boxes) in oriC, the replication origin. DnaA is an ATP-binding protein and its binding to DNA is affected depending on whether ATP, ADP, or no nucleotide is bound. One mechanism by which the activity of the replication origin is controlled is DNA methylation. The E. coli oriC contains 11 copies of the sequence, which is a target for methylation at the N position of adenine by the Dam methylase enzyme. These sites are also found scattered throughout the genome. Note, though, that several of these methylation sites overlap dnaA boxes, as illustrated in FIGURE 1.

FIGURE 1. The E. coli origin of replication, oriC, contains multiple binding sites for the DnaA initiator protein. In a number of cases these sites overlap Dam methylation sites.
Before replication, the palindromic target site is methylated on the adenines of each strand. Replication inserts the normal (nonmodified) bases into the daughter strands. This generates hemimethylated DNA, in which one strand is methylated and one strand is unmethylated. Thus, the replication event converts Dam target sites from fully methylated to hemimethylated condition.
What is the consequence for replication? The ability of a plasmid relying upon oriC to replicate in dam E. coli depends on its state of methylation. If the plasmid is methylated, it undergoes a single round of replication, and then the hemimethylated products accumulate, as described in FIGURE 2. The hemimethylated plasmids then accumulate rather than being replaced by unmethylated plasmids, suggesting that a hemimethylated origin cannot be used to initiate a replication cycle.
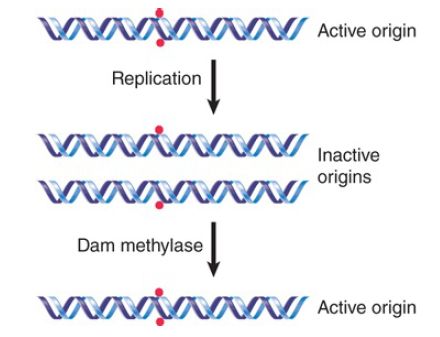
FIGURE 2. Only fully methylated origins can initiate replication; hemimethylated daughter origins cannot be used again until they have been restored to the fully methylated state.
This suggests two explanations: Initiation might require full methylation of the Dam target sites in the origin, or it might be inhibited by hemimethylation of these sites. The latter seems to be the case, because an origin of nonmethylated DNA can function effectively.
Thus hemimethylated origins cannot initiate again until the Dam methylase has converted them into fully methylated origins. The GATC sites at the origin remain hemimethylated for approximately 13 minutes after replication. This long period is unusual because at typical GATC sites elsewhere in the genome, remethylation begins immediately (less than 1.5 minutes) following replication. One other region behaves like oriC: The promoter of the dnaA gene also shows a delay before remethylation begins. Even though it is hemimethylated, the dnaA gene promoter is repressed, which causes a reduction in the level of DnaA protein. Thus, the origin itself is inert, and production of the crucial initiator protein is repressed during this period.
DNA methylation in bacteria serves a second function, as well: It allows the DNA mismatch recognition machinery to distinguish the old template strand from the new strand. If the DNA polymerase has made an error, such as creating an A-C base pair, the repair system will use the methylated strand as a template to replace the base on the nonmethylated strand. Without that methylation, the enzyme would have no way to determine which is the new strand.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)