النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Replication of Chromatin Requires Assembly of Nucleosomes
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
25-3-2021
3185
Replication of Chromatin Requires Assembly of Nucleosomes
KEY CONCEPTS
-Histone octamers are not conserved during replication, but H2A-H2B dimers and H32 -H42 tetramers are.
-There are different pathways for the assembly of nucleosomes during replication and also independent of replication.
-Accessory proteins are required to assist the assembly of nucleosomes.
-CAF-1 and ASF1 are histone assembly proteins that are linked to the replication machinery.
-A different assembly protein, HIRA, and the histone H3.3 variant are used for replication-independent assembly.
Replication separates the strands of DNA and therefore must inevitably disrupt the structure of the nucleosome. However, this disruption is confined to the immediate vicinity of the replication fork. As soon as DNA has been replicated, nucleosomes are quickly generated on both of the duplicates. The transience of the replication event is a major difficulty in analyzing the structure of a particular region while it is being replicated.
Replication of chromatin does not involve any protracted period during which the DNA is free of histones. This point is illustrated by the electron micrograph of FIGURE 1, which shows a recentl replicated stretch of DNA that is already packaged into nucleosomes on both daughter duplex segments.
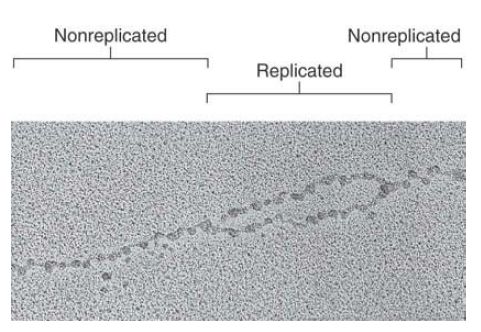
FIGURE 1 Replicated DNA is immediately incorporated into nucleosomes.
Photo courtesy of Steven L. McKnight, UT Southwestern Medical Center at Dallas.
Biochemical analysis and visualization of the replication fork indicate that the disruption of nucleosome structure is limited to a short region immediately around the fork. Progress of the fork disrupts nucleosomes, but they form very rapidly on the daughter duplexes as the fork moves forward. In fact, the assembly of nucleosomes is directly linked to the replisome that is replicating DNA.
How do histones associate with DNA to generate nucleosomes? Do the histones preform a protein octamer around which the DNA is subsequently wrapped? Or, does the histone octamer assemble on DNA from free histones? Researchers can use either of these pathways in vitro to assemble nucleosomes, depending on the conditions that are employed. In one pathway, a preformed octamer binds to DNA. In the other pathway, a tetramer of H32 -H42 binds first, and then two H2A-H2B dimers are added. This latter stepwise assembly is the pathway that is used in replication, shown in FIGURE 2.

FIGURE 2 During nucleosome assembly in vivo, H3-H4 tetramers form and bind DNA first, then two H2A-H2B dimers are added to form the complete nucleosome.
Accessory proteins are involved in assisting histones to associate with DNA. Accessory proteins can act as “molecular chaperones” that bind to the histones in order to release either individual histones or complexes (H32 -H42 or H2A-H2B) to the DNA in a controlled manner. This could be necessary because the histones, as basic proteins, have a generally high affinity for DNA. Such interactions allow histones to form nucleosomes without becoming trapped in other kinetic intermediates (i.e., other complexes resulting from indiscreet binding of histones to DNA).
Researchers have identified numerous histone chaperones. Chromatin assembly factor (CAF)-1 and anti-silencing function 1 (ASF1) are two chaperones that function at the replication fork. CAF-1 is a conserved three-subunit complex that is directly recruited to the replication fork by proliferating cell nuclear antigen (PCNA), the processivity factor for DNA polymerase. ASF1 interacts with the replicative helicase that unwinds the replication fork. Furthermore, CAF-1 and ASF1 interact with each other.
These interactions provide the link between replication and nucleosome assembly, ensuring that nucleosomes are assembled as soon as DNA has been replicated.
CAF-1 acts stoichiometrically, and functions by binding to newly synthesized H3 and H4. New nucleosomes form by assembling first the H32 -H42 tetramer, and then adding the H2A-H2B dimers. ASF1 appears to play an important role in transfer of parental nucleosomes from ahead of the replication fork to the newly synthesized region behind the fork, although ASF1 can bind and assemble newly synthesized histones, as well.
The pattern of disassembly and reassembly has been difficult to characterize in detail, but a working model is illustrated in FIGURE 3. The replication fork displaces histone octamers, which then dissociate into H32 -H42 tetramers and H2A-H2B dimers. These “old” tetramers and dimers enter a pool that also includes “new” tetramers and dimers, which are assembled from newly synthesized histones. Nucleosomes assemble ~600 bp behind th ereplication fork. Assembly is initiated when H32 -H42 tetramers bind to each of the daughter duplexes, assisted by CAF-1 or ASF1. Two H2A-H2B dimers then bind to each H32 -H42 tetramer to complete the histone octamer. The assembly of tetramers and dimers is random with respect to “old” and “new” subunits. It appears that nucleosomes are disrupted and reassembled in a similar way during transcription, though different histone chaperones are involved in this process .

FIGURE 2 Replication fork passage displaces histone octamers from DNA. They disassemble into H3-H4 tetramers and H2A-H2B dimers. H3-H4 tetramers (blue) are directly transferred behind the replication forks. Newly synthesized histones (orange) are assembled into H3-H4 tetramers and H2A-H2B dimers. The old and new tetramers and dimers are assembled with the aid of histone chaperones into new nucleosomes immediately behind the replication fork. H2A-H2B dimers are omitted from the figure for simplicity; chaperones responsible for dimer assembly have not been identified.
Data from: Rocha, W., and Verreault, A. 2008. FEBS Lett 582:1938–1949.
During S phase (the period of DNA replication) in a eukaryotic cell, the duplication of chromatin requires synthesis of sufficient histone proteins to package an entire genome—basically the same quantity of histones must be synthesized that are already contained in nucleosomes. The synthesis of histone mRNAs is controlled as part of the cell cycle, and increases enormously in S phase. The pathway for assembling chromatin from this equal mix of old and new histones during S phase is called the replication-coupled pathway.
Another pathway, called the replication-independent pathway, exists for assembling nucleosomes during other phases of the cell cycle, when DNA is not being synthesized. This might become necessary as the result of damage to DNA or because nucleosomes are displaced during transcription. The assembly process must necessarily have some differences from the replication-coupled pathway, because it cannot be linked to the replication apparatus. The replication-independent pathway uses the histone H3.3 variant, which was introduced earlier in the section Histone Variants Produce Alternative Nucleosomes.
The histone H3.3 variant differs from the highly conserved H3 histone at four amino acid positions . H3.3 slowly replaces H3 in differentiating cells that do not have replication cycles. This happens as the result of assembly of new histoneoctamer s to replace those that have been displaced from DNA for whatever reason. The mechanism that is used to ensure the use of H3.3 in the replication-independent pathway is different in two cases that have been investigated.
In the protozoan Tetrahymena, histone usage is determined exclusively by availability. Histone H3 is synthesized only during the cell cycle; the variant replacement histone is synthesized only in nonreplicating cells. In Drosophila, however, there is an active pathway that ensures the usage of H3.3 by the replicationindependent pathway. New nucleosomes containing H3.3 assemble at sites of transcription, presumably replacing nucleosomes that were displaced by RNA polymerase. The assembly process discriminates between H3 and H3.3 on the basis of their sequences, specifically excluding H3 from being utilized. By contrast, replication-coupled assembly uses both types of H3 (although H3.3 is available at much lower levels than H3 and
therefore enters only a small proportion of nucleosomes).
CAF-1 is not involved in replication-independent assembly. (There also are organisms such as yeast and Arabidopsis for which its gene is not essential, implying that alternative assembly processes can be used in replication-coupled assembly.) Instead, replicationindependent assembly uses a factor called HIRA, named for histone cell cycle regulator (HIR), genes in yeast. Depletion of HIRA from in vitro systems for nucleosome assembly inhibits the formation of nucleosomes on nonreplicated DNA, but not on replicating DNA, which indicates that the pathways do indeed use different assembly mechanisms. Like CAF-1 and ASF1, HIRA
functions as a chaperone to assist the incorporation of histones into nucleosomes. This pathway appears to be generally responsible for replication-independent assembly; for example, HIRA is required for the decondensation of the sperm nucleus, when protamines are replaced by histones, in order to generate chromatin that is competent to be replicated following fertilization.
As described earlier, assembly of nucleosomes containing an alternative to H3 also occurs at centromeres . Centromeric DNA replicates early during S phase. The incorporation of H3 at the centromeres is inhibited during replication; instead, a CenH3 variant is preferentially (though not exclusively) incorporated. Interestingly, new CenH3 is incorporated during early G1 in vertebrates, but in budding yeast the CenH3 is incorporated in S phase and is linked to replication. In both vertebrates and yeast, CenH3 incorporation requires a CenH3-specific chaperone, called HJURP (mammals) or Scm3 (budding yeast).
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)