


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية| Eukaryotic Genomes Contain Nonrepetitive and Repetitive DNA Sequences |
|
|
|
Read More
Date: 8-12-2015
Date: 7-11-2020
Date: 25-5-2021
|
Eukaryotic Genomes Contain Nonrepetitive and Repetitive DNA Sequences
KEY CONCEPTS
- The kinetics of DNA reassociation after a genome has been denatured distinguish sequences by their frequency of repetition in the genome.
- Polypeptides are generally encoded by sequences in nonrepetitive DNA.
- Larger genomes within a taxonomic group do not contain more genes but have large amounts of repetitive DNA.
- A large part of moderately repetitive DNA can be made up of transposons.
The general nature of the eukaryotic genome can be assessed by the kinetics of reassociation of denatured DNA. Researchers used this technique extensively before large-scale DNA sequencing became possible.
Reassociation kinetics identifies two general types of genomic sequences:
- Nonrepetitive DNA consists of sequences that are unique: there is only one copy in a haploid genome.
- Repetitive DNA consists of sequences that are present in more than one copy in each haploid genome.
We can divide repetitive DNA into two general types:
- Moderately repetitive DNA consists of relatively short sequences that are repeated typically 10 to 1,000 times in the genome. The sequences are dispersed throughout the genome and are responsible for the high degree of secondary structure formation in pre-mRNA when inverted repeats in the introns pair to form duplex regions. Genes for tRNAs and rRNAs are also moderately repetitive.
- Highly repetitive DNA consists of very short sequences (typically fewer than 100 base pairs [bp]) that are present many thousands of times in the genome, often organized as long regions of tandem repeats (see the Clusters and Repeats chapter). Neither class is found in exons.
The proportion of the genome occupied by nonrepetitive DNA varies widely among taxonomic groups. FIGURE 1 summarizes the genome organization of some representative organisms. Prokaryotes contain nonrepetitive DNA almost exclusively. For unicellular eukaryotes, most of the DNA is nonrepetitive: less than 20% fall into one or more moderately repetitive components. In animal cells, up to half of the DNA is represented by moderately and highly repetitive components. In plants and amphibians, the moderately and highly repetitive components can account for up to80% of the genome, so that the nonrepetitive DNA is reduced to a small component.
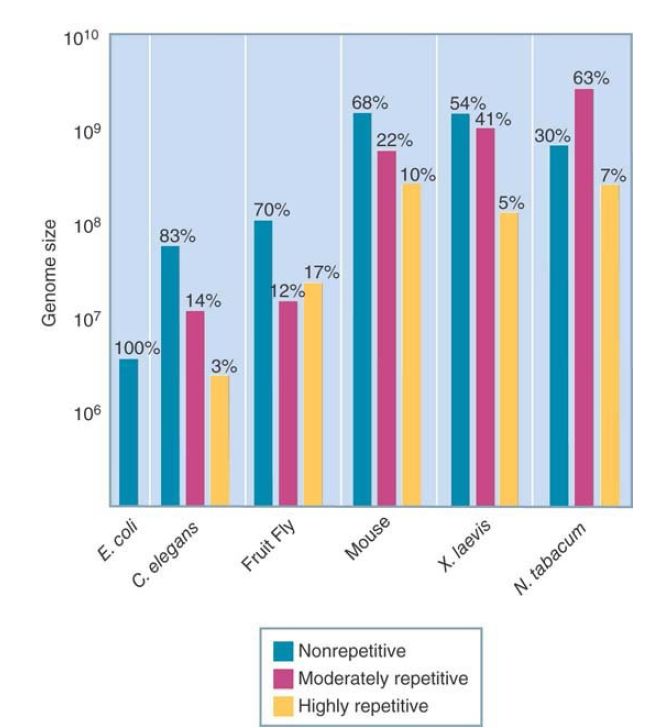
FIGURE 1. The proportions of different sequence components vary in eukaryotic genomes. The absolute content of nonrepetitive DNA increases with genome size but reaches a plateau at about 2 × 109 bp.
A significant part of the moderately repetitive DNA consists of transposons, short sequences of DNA (up to about 5 kilobases [kb]) that have the ability to move to new locations in the genome and/or to make additional copies of themselves . In some multicellular eukaryotic genomes they may even occupy more than half of the genome .
Transposons were historically viewed as selfish DNA, which is defined as sequences that propagate themselves within a genome without contributing to the development and functioning of the organism. Transposons are not necessarily “selfish,” because they can cause genome rearrangements, which could confer selective advantages. It is fair to say, though, that we do not really understand why selective forces do not act against transposons becoming such a large proportion of the eukaryotic genome. It might be that they are selectively neutral as long as they do not interrupt or delete coding or regulatory regions. Many organisms have active cellular transposition suppression mechanisms, perhaps because in some cases deleterious chromosome breakages result.
Another term used to describe the apparent excess of DNA in some genomes is junk DNA, meaning genomic sequences without any apparent function, though this name might simply reflect our failure to understand the functions of many of these sequences. Of course, it is likely that there is a balance in the genome between the generation of new sequences and the elimination of unneededsequences, and som e proportion of DNA that apparently lacks function might be destined to be eliminated.
The length of the nonrepetitive DNA component tends to increase with overall genome size up to a total genome size of about 3 × 109 bp (characteristic of mammals). However, further increases in genome size generally reflect an increase in the amount and proportion of the repetitive components, so that it is rare for an organism to have a nonrepetitive DNA component greater than 2 × 109 bp. Therefore, the nonrepetitive DNA content of genomes is a
better indication of the relative complexity of the organism.
Escherichia coli (a prokaryote) has 4.2 × 106 bp of nonrepetitive DNA; Caenorhabditis elegans (a multicellular eukaryote) has an order of magnitude more at 6.6 × 107 bp; Drosophila melanogaster has about 10 bp; and mammals have yet another order of magnitude more, at about 2 × 109 bp.
What type of DNA corresponds to polypeptide-coding genes? Reassociation kinetics typically shows that mRNA is transcribed from nonrepetitive DNA. Therefore, the amount of nonrepetitive DNA is a better indication of the coding potential than is the size of the genome. (However, more detailed analysis based on genomic sequences shows that many exons have related sequences in other exons . Su ch exonsevolve by duplication to result in copies that initially are identical but that then diverge in sequence during evolution.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
اتحاد كليات الطب الملكية البريطانية يشيد بالمستوى العلمي لطلبة جامعة العميد وبيئتها التعليمية
|
|
|