


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 13-12-2015
Date: 24-12-2015
Date: 21-3-2021
|
Some DNA Sequences Encode More Than One Polypeptide
KEY CONCEPTS
-The use of alternative initiation or termination codons allows multiple variants of a polypeptide chain.
-Different polypeptides can be produced from the same sequence of DNA when the mRNA is read in different reading frames (as two overlapping genes).
-Otherwise identical polypeptides, differing by the presence or absence of certain regions, can be generated by differential (alternative) splicing. This can take the form of including or excluding individual exons, or of choosing between alternative exons.
Many structural genes consist of a sequence that encodes a single polypeptide, although the gene can include noncoding regions at both ends and introns within the coding region. However, there are some cases in which a single sequence of DNA encodes more than one polypeptide.
In one simple example, a single DNA sequence can have two alternative start codons in the same reading frame (see FIGURE 1). Thus, under different conditions one or the other of the start codons might be used, allowing the production of either a short form of the polypeptide or a full-length form, where the short form is the last portion of the full-length form.
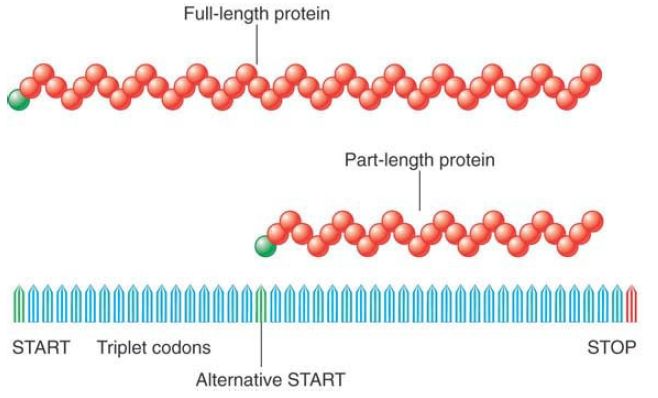
FIGURE 1.Two proteins can be generated from a single gene by starting (or terminating) expression at different points.
An actual overlapping gene occurs when the same sequence of DNA encodes two nonhomologous proteins because it uses more than one reading frame. Usually, a coding DNA sequence is read in only one of the three potential reading frames. In some viral and mitochondrial genes, however, there is some overlap between two adjacent genes that are read in different reading frames, as illustrated in FIGURE2. The length of overlap is usually short, so that most of the DNA sequence encodes a unique polypeptide sequence.

FIGURE 2 Two genes might overlap by reading the same DNA sequence in different frames.
In some cases, genes can be nested. This occurs when a complete gene is found within the intron of a larger “host” gene. Nested genes often lie on the strand opposite that of the host gene. In some genes there are switches in the pathway for splicing the exons that result in alternative patterns of gene expression. A single gene might generate a variety of mRNA products that differ in their exon content. Certain exons might be optional; in other words, they might be included or spliced out. There also might be a pair of exons treated as mutually exclusive—one or the other is included in the mature transcript, but not both. The alternative proteins have one part in common and one unique part.
In some cases, the alternative means of expression do not affect the sequence of the polypeptide. For example, changes that affect the 5′ UTR or the 3′ UTR might have regulatory consequences, but the same polypeptide is made. In other cases, one exon is substituted for another, as in FIGURE 3. In this example, the polypeptides produced by the two mRNAs contain sequences that overlap extensively, but are different within the alternatively spliced region. The 3′ half of the troponin T gene of rat muscle contains five exons, but only four are used to construct an individual mRNA. Three exons (W, X, and Z) are included in all mRNAs. However, in one alternative splicing pattern, the α exon is included between X and Z, whereas in the other pattern it is replaced by the β exon.
The α and β forms of troponin T therefore differ in the sequence of the amino acids between W and Z, depending on which of the alternative exons (α or β) is used. Either one of the α and β exons can be used in an individual mRNA, but both cannot be used in the same mRNA.
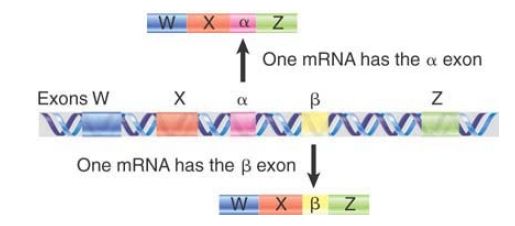
FIGURE 3.13 Alternative splicing generates the a and b variants of troponin T.
FIGURE 4 shows that alternative splicing can lead to the inclusion of an exon in some mRNAs, whereas it leaves it out of others. A single primary transcript can be spliced in either of two ways. In the first (more standard) pathway, two introns are spliced out and the three exons are joined together. In the second pathway, the second exon is excluded as if a single large intron is spliced out. This intron consists of intron 1 + exon 2 + intron 2. In effect, exon 2 has been treated in this pathway as if it were part of a single intron. The pathways produce two polypeptides that are the same at their ends, but one has an additional sequence in the middle.
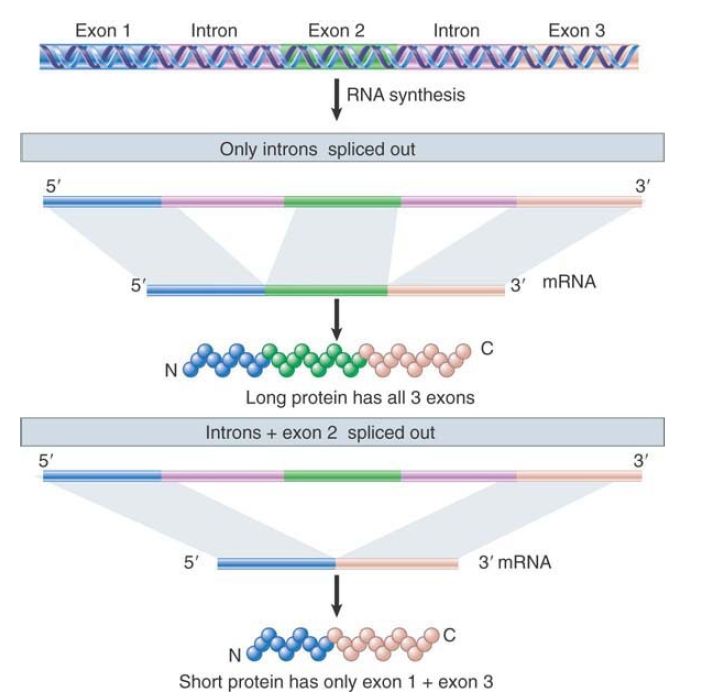
FIGURE 4. Alternative splicing uses the same pre-mRNA to generate mRNAs that have different combinations of exons.
Sometimes two alternative splicing pathways operate simultaneously, with a certain proportion of the primary RNA transcripts being spliced in each way. However, sometimes the pathways are alternatives that are expressed under different conditions; for example, one in one cell type and one in another cell type.
So, alternative (or differential) splicing can generate different polypeptides with related sequences from a single stretch of DNA. It is curious that the multicellular eukaryotic genome is often extremely large with long genes that are often widely dispersed along a chromosome, but at the same time there might be multiple products from a single locus. Due to alternative splicing, there are about 15% more polypeptides than genes in flies and worms, but it is estimated that the majority of human genes are alternatively spliced (see the chapter titled Genome Sequences and Evolution).



|
|
|
|
مخاطر خفية لمكون شائع في مشروبات الطاقة والمكملات الغذائية
|
|
|
|
|
|
|
"آبل" تشغّل نظامها الجديد للذكاء الاصطناعي على أجهزتها
|
|
|
|
|
|
|
المجمع العلميّ يُواصل عقد جلسات تعليميّة في فنون الإقراء لطلبة العلوم الدينيّة في النجف الأشرف
|
|
|