
Hierarchical Sequencing
 المؤلف:
John M Walker and Ralph Rapley
المؤلف:
John M Walker and Ralph Rapley
 المصدر:
Molecular Biology and Biotechnology 5th Edition
المصدر:
Molecular Biology and Biotechnology 5th Edition
 الجزء والصفحة:
الجزء والصفحة:
 27-11-2020
27-11-2020
 2489
2489
Hierarchical Sequencing
This approach relies on the production of a set of large-insert clones (typically 100–200 kb each) that cover the entire genome.Since the clones are all independent and ultimately positioned on a physical map in an order that represents each chromosome, repeated sequences are far less troublesome, leading to fewer gaps than encountered by the WGS approach. For many genome projects, high-capacity vectors such as BACs are advantageously used for generating large-insert clones since they are less likely to rearrange than alternatives such as YACs. Longrange physical maps are generated from the production of ‘contigs’.
Contigs are a set of overlapping DNA fragments that have been obtained from independent clones and positioned relative to one another so that they form a contiguous array. To obtain contigs, genomic libraries must prepared from high molecular weight DNA that has either been partially digested with restriction enzymes or randomly sheared. Partial digestion or random shearing leads to the production of a set of overlapping clones, whereas complete digestion would produce a set of fragments with no overlaps (Figure 1). Partial digestion ensures that when each DNA fragment is cloned into a vector, it has ends that will overlap with other clones. Thus, when the overlaps are identified, the clones can be positioned or ordered, so that a physical map is produced.

Figure 1 Comparison of partial and complete digestion of DNA molecules at restriction enzymes sites (E).
Large-insert clones are broken down further into sets of smaller overlapping subclones and sequenced using the shotgun sequencing approach. In order to position the overlapping ends into a contig representing the large insert clone, it is preferable to undertake DNA sequencing of both ends of the individual subclones (double-barrelled
shotgun sequencing). Eventually, the entire DNA sequence of the largeinsert clone is obtained by computer-based alignment of individual subclone DNA end sequences. In order to minimise the overlaps and identify the large-insert clones for further sequencing, restriction enzyme mapping can be undertaken to produce a ‘fingerprint clone contig’. In the Human Genome Project, fingerprint clone contigs were mapped to
human chromosomal locations by each chromosome workgroup using resources such as panels of human radiation hybrids (RH), fluorescence in situ hybridisation (FISH) with human chromosomes and existing genetic maps. Radiation hybrids are panels of human–hamster cell hybrids formed by fusing human cells containing radiation-generated fragments of human chromosomes with hamster cells. Panels of radiation hybrids that contain characterized fragments from all human chromosomes can be used for constructing genetic maps that are complementary to both recombination maps and physical maps based on contigs. In order to define a common way for all research laboratories to order clones and connect physical maps together, an arbitrary molecular technique based on the PCR has been developed to generate sequencetagged sites (STS). These are small, unique sequences between 200 and 300 base pairs that are amplified by PCR.80 The uniqueness of the STS is defined by the PCR primers that flank the STS. If the PCR results in amplification, then the STS is present in the clone being tested. In this way, defining STS markers that lie approximately 100 kb apart along a contig map allows the ordering of those contigs. Thus, all groups working with clones have publicly available defined landmarks with which to order clones produced in their DNA libraries (Figure 2).
STSs may also be generated from polymorphic markers that may be traced through families along with other DNA markers and located on a genetic linkage map. These polymorphic STSs may thus serve as markers on both a physical map and a genetic linkage map for each chromosome and therefore provide a useful means for aligning the two types of map.
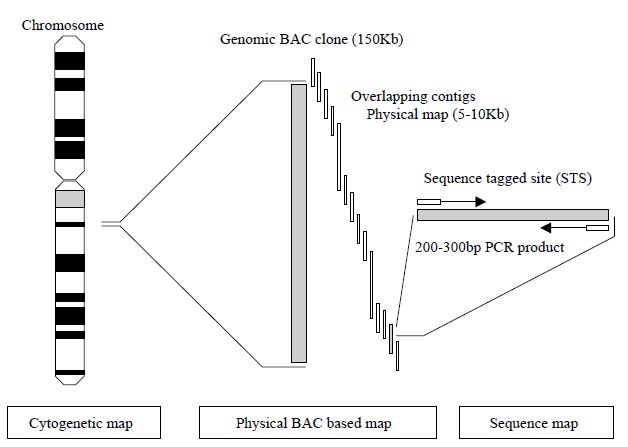
Figure 2. Schematic of the use of STS markers in the hierarchical physical mapping
a human chromosome using BAC clones.
In addition to the human genome, the hierarchical sequencing approach has been used to sequence several genomes, including those of the yeast Saccharomyces cerevisiae and the nematode worm Cae- norhabditis elegans.High-quality BAC clone-based physical genome maps in one species can be of great value to genome projects in other species where some conservation of genomic sequence and gene order might be expected. For example, the outputs of the Human Genome Project have provided anchors for ordering BAC clones generated in several other species such as mouse, rat and cattle, allowing simplification of clone alignments and physical map building in addition to the generation of comparative maps in the respective species. The International Human Genome Sequencing Consortium sequence was reported as finished in 2004 (Build 35) and contains 2.85 billion nucleotides interrupted by 341 gaps. It covers approximately 99% of the euchromatic genome.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


