


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 25-12-2015
Date: 29-5-2021
Date: 30-4-2021
|
Prokaryotic mRNA Degradation Involves Multiple Enzymes
KEY CONCEPTS
- Degradation of bacterial mRNAs is initiated by removal of a pyrophosphate from the 5′ terminus.
- Monophosphorylated mRNAs are degraded during translation in a two-step cycle involving endonucleolytic cleavages, followed by 3′ to 5′ digestion of the resulting fragments.
- 3′ polyadenylation can facilitate the degradation of mRNA fragments containing secondary structure.
- The main degradation enzymes work as a complex called the degradosome.
Our understanding of prokaryotic mRNA degradation comes mostly from studies of E. coli. So far, the general principles apply to the other bacterial species studied. In prokaryotes, mRNA degradation occurs during the process of coupled transcription/translation.
Prokaryotic ribosomes begin translation even before transcription is completed, attaching to the mRNA at an initiation site near the 5′ end and proceeding toward the 3′ end. Multiple ribosomes can initiate translation on the same mRNA sequentially, forming a polyribosome (or polysome): one mRNA with multiple ribosomes.
E. coli mRNAs are degraded by a combination of endonuclease and 3′ to 5′ exonuclease activities. The major mRNA degradation pathway in E. coli is a multistage process illustrated in FIGURE 1. The initiating step is removal of pyrophosphate from the 5′ terminus, leaving a single phosphate. The monophosphorylated form stimulates the catalytic activity of an endonuclease (RNase E), which makes an initial cut near the 5′ end of the mRNA. This cleavage leaves a 3′–OH on the upstream fragment and a 5′–monophosphate on the downstream fragment. It functionally destroys a monocistronic mRNA, because ribosomes can no longer initiate translation. The upstream fragment is then degraded by a 3′ to 5′ exonuclease (polynucleotide phosphorylase, or PNPase). This two-step ribonuclease cycle is repeated along the length of the mRNA in a 5′ to 3′ direction as more RNA gets exposed following passage of previously initiated ribosomes. This process proceeds very rapidly as the short fragments generated by RNase E can be detected only in mutant cells in which exonuclease activity is impaired.
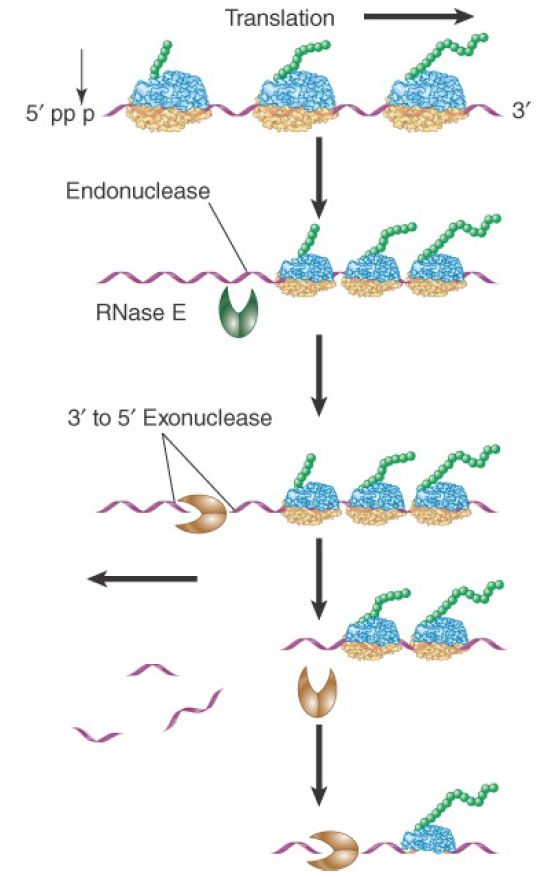
FIGURE 1. Degradation of bacterial mRNAs. Bacterial mRNA degradation is initiated by cleavage of the triphosphate 5′ terminus to yield a monophosphate. mRNAs are then degraded in a twostep cycle: an endonucleolytic cleavage, followed by 3′ to 5′ exonuclease digestion of the released fragment. The endonucleolytic cleavages occur in a 5′ to 3′ direction on the mRNA, following the passage of the last ribosome.
PNPase, as well as the other known 3′ to 5′ exonucleases in E.coli, are unable to progress through double-stranded regions. Thus, the stem-loop structure at the 3′ end of many bacterial mRNAs protects the mRNA from direct 3′ attack. Some internal fragments generated by RNase E cleavage also have regions of secondary structure that would impede exonuclease digestion.
PNPase is, however, able to digest through double-stranded regions if there is a stretch of single-stranded RNA at least 7 to 10 nucleotides long located 3′ to the stem-loop. The single-stranded sequence seems to serve as a necessary staging platform for the enzyme. Rho-independent termination leaves a single-stranded region that is too short to serve as a platform. To solve this problem a bacterial poly (A) polymerase (PAP) adds 10 to 40 nucleotide poly(A) tails to 3′ termini, making them susceptible to 3′ to 5′ degradation. RNA fragments terminating in particularly stable secondary structures may require repeated polyadenylation and exonuclease digestion steps. It is not known whether polyadenylation is ever the initiating step for degradation of mRNA, or whether it is used only to help degrade fragments, including the 3′ terminal one. Some experiments indicate that RNase E cleavage of an mRNA may be required to activate the PAP. This would explain why intact mRNAs do not seem to be degraded from the 3′ end.
RNase E and PNPase, along with a helicase and another accessory enzyme, form a multiprotein complex called the degradosome. RNase E plays dual roles in the complex. Its Nterminal domain provides the endonuclease activity, whereas its Cterminal domain provides a scaffold that holds together the other components. Although RNase E and PNPase are the principal endo- and exonucleases active in mRNA degradation, others also exist, probably with more restricted roles. The role of other nucleases in mRNA degradation has been addressed by evaluating the phenotypes of mutants in each of the enzymes. For example, the inactivation of RNase E slows mRNA degradation without completely blocking it. Mutations that inactivate PNPase or either of the other two known 3′ to 5′ exonucleases have essentially no effect on overall mRNA stability. This reveals that any pair of the exonucleases can carry out apparently normal mRNA degradation.
However, only two of the three exonucleases (PNPase and RNase R) can digest fragments with stable secondary structures. This was demonstrated in double-mutant studies, in which both PNPase and RNase R are inactivated. mRNA fragments that contain secondary structures accumulated in these mutants.
Many questions about mRNA degradation in E. coli remain to be answered. Half-lives for different mRNAs in E. coli can differ more than 100-fold. The basis for these extreme differences in stability is not fully understood but appears to be largely due to two factors. Different mRNAs exhibit a range of susceptibilities to endonuclease cleavage, with some protection being conferred by the secondary structure of the 5′ end region. Some mRNAs are more efficiently translated than others, resulting in a denser packing of protective ribosomes. Whether or not there are additional pathways of mRNA degradation is not known. No 5′ to 3′ exonuclease has been found in E. coli, though one has been identified in Bacillus subtilis and some other bacterial species. So far, the bacterial species found to have the 5′ to 3′ exonuclease RNase J lack the endonuclease RNase E (the major degradative RNase in E. coli). This suggests there is at least one alternative mRNA decay pathway in bacteria.
It is likely that the different endonucleases and exonucleases have distinct roles. A genome-wide study using microarrays looked at the steady-state levels of more than 4,000 mRNAs in cells mutant for RNase E or PNPase or other degradosome components. Many mRNA levels increased in the mutants, as expected for a decrease in degradation. Others, however, remained at the same level or even decreased. The half-lives of specific mRNAs can be altered by different cellular physiological states such as starvation or other forms of stress, and mechanisms for these changes remain mostly unknown.



|
|
|
|
التوتر والسرطان.. علماء يحذرون من "صلة خطيرة"
|
|
|
|
|
|
|
مرآة السيارة: مدى دقة عكسها للصورة الصحيحة
|
|
|
|
|
|
|
مجلةٌ عالميةٌ تنشر تفاصيل مشروع ملف تفويض المؤلِّفين العراقيين لحفظ التراث الفكري
|
|
|