


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية| About 10,000 Genes Are Expressed at Widely Differing Levels in a Eukaryotic Cell |
|
|
|
Read More
Date: 17-11-2020
Date: 3-1-2016
Date: 28-10-2020
|
About 10,000 Genes Are Expressed at Widely Differing Levels in a Eukaryotic Cell
KEY CONCEPTS
-In any particular cell, most genes are expressed at a low level.
-Only a small number of genes, whose products are specialized for the cell type, are highly expressed.
-mRNAs expressed at low levels overlap extensively when different cell types are compared.
-The abundantly expressed mRNAs are usually specific for the cell type.
-About 10,000 expressed genes might be common to most cell types of a multicellular eukaryote.
The proportion of DNA containing protein-coding genes being expressed in a specific cell at a specific time can be determined by the amount of the DNA that can hybridize with the mRNAs isolated from that cell. Such a saturation analysis conducted for many cell types at various times typically identifies about 1% of the DNA being expressed as mRNA. From this researchers can calculate the number of protein-coding genes, as long as they know the average length of an mRNA. For a unicellular eukaryote such as yeast, the total number of expressed protein-coding genes is about 4,000. For somatic tissues of multicellular eukaryotes, including both plants and vertebrates, the number is usually 10,000 to 15,000. (The only consistent exception to this type of value is presented by mammalian brain cells, for which much larger numbers of genes appear to be expressed, although the exact number is not certain.)
Researchers can use kinetic analysis of the reassociation of an RNA population to determine its sequence complexity. This type of analysis typically identifies three components in a eukaryotic cell.
Just as with a DNA reassociation curve, a single component hybridizes over about 2 decades of Rot values (RNA concentration × time), and a reaction extending over a greater range must be resolved by computer curve-fitting into individual components.
Again, this represents what is really a continuous spectrum of sequences. FIGURE 1. shows an example of an excess mRNA × cDNA reaction that generates three components:
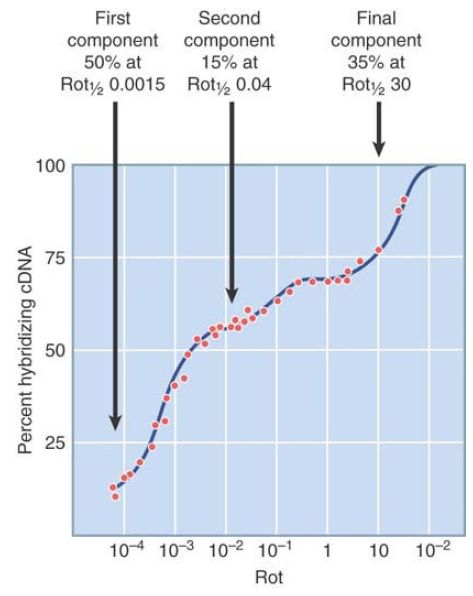
FIGURE 1. Hybridization between excess mRNA and cDNA identifies several components in chick oviduct cells, each characterized by the Rot 1/2 of reaction.
- The first component has the same characteristics as a control reaction of ovalbumin mRNA with its DNA copy. This suggests that the first component is in fact just ovalbumin mRNA (which indeed is about half of the mRNA mass in oviduct tissue).
- The next component provides 15% of the reaction, with a total length of 15 kb. This corresponds to 7 to 8 mRNA species with an average length of 2,000 bases.
- The last component provides 35% of the reaction, which corresponds to a length of 26 Mb. This corresponds to about 13,000 mRNA species with an average length of 2,000 bases.
From this analysis, we can see that about half of the mass of mRNA in the cell represents a single mRNA, about 15% of the mass is provided by a mere seven to eight mRNAs, and about 35% of the mass is divided into the large number of 13,000 mRNA types. It is therefore obvious that the mRNAs comprising each component must be present in very different amounts.
The average number of molecules of each mRNA per cell is called its abundance. Researchers can calculate it quite simply if the total mass of a specific mRNA type in the cell is known. In the example of chick oviduct cells shown in Figure 5.17, the total mRNA can be accounted for as 100,000 copies of the first component (ovalbumin mRNA), 4,000 copies of each of 7 or 8 other mRNAs in the second component, and only about 5 copies of each of the 13,000 remaining mRNAs that constitute the last component.
We can divide the mRNA population into two general classes, according to their abundance:
- The oviduct is an extreme case, with so much of the mRNA represented by only one type, but most cells do contain a small number of RNAs present in many copies each. This abundant mRNA component typically consists of fewer than 100 different mRNAs present in 1,000 to 10,000 copies per cell. It often corresponds to a major part of the mass, approaching 50% of the total mRNA.
- About half of the mass of the mRNA consists of a large number of sequences, of the order of 10,000, each represented by only a small number of copies in the mRNA—say, fewer than 10. This is the scarce mRNA (or complex mRNA) class. It is this class that drives a saturation reaction.
Many somatic tissues of multicellular eukaryotes have an expressed gene number in the range of 10,000 to 20,000. How much overlap is there between the genes expressed in different tissues? For example, the expressed gene number of chick liver is between 11,000 and 17,000, compared with the value for oviduct of 13,000 to 15,000. How much do these two sets of genes
overlap? How many are specific for each tissue? These questions are usually addressed by analyzing the transcriptome—the set of sequences represented in RNA.
We see immediately that there are likely to be substantial differences among the genes expressed in the abundant class. Ovalbumin, for example, is synthesized only in the oviduct and not at all in the liver. This means that 50% of the mass of mRNA in the oviduct is specific to that tissue.
However, the abundant mRNAs represent only a small proportion of the number of expressed genes. In terms of the total number of genes of the organism, and of the number of changes in transcription that must be made between different cell types, we need to know the extent of overlap between the genes represented in the scarce mRNA classes of different cell phenotypes.
Comparisons between different tissues show that, for example, about 75% of the sequences expressed in liver and oviduct are the same. In other words, about 12,000 genes are expressed in both liver and oviduct, 5,000 additional genes are expressed only in liver, and 3,000 additional genes are expressed only in oviduct.
The scarce mRNAs overlap extensively. Between mouse liver and kidney, about 90% of the scarce mRNAs are identical, leaving a difference between the tissues of only 1,000 to 2,000 expressed genes. The general result obtained in several comparisons of this sort is that only about 10% of the mRNA sequences of a cell are unique to it. The majority of mRNAs are common to many—perhaps even all—cell types.
This suggests that the common set of expressed gene functions, numbering perhaps about 10,000 in mammals, comprise functions that are needed in all cell types. Sometimes, this type of function is referred to as a housekeeping gene or constitutive gene. Itcont rasts with the activities represented by specialized functions (such as ovalbumin or globin) needed only for particular cell phenotypes. These are sometimes called luxury genes.



|
|
|
|
دخلت غرفة فنسيت ماذا تريد من داخلها.. خبير يفسر الحالة
|
|
|
|
|
|
|
ثورة طبية.. ابتكار أصغر جهاز لتنظيم ضربات القلب في العالم
|
|
|
|
|
|
|
العتبة العباسية المقدسة تستعد لإطلاق الحفل المركزي لتخرج طلبة الجامعات العراقية
|
|
|